6VYH
 
 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
6WIK
 
 | | Cryo-EM structure of SLC40/ferroportin with Fab in the presence of hepcidin | | Descriptor: | 11F9 Fab heavy-chain, 11F9 Fab light-chain, Solute carrier family 40 protein | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-11 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
3IFI
 
 | | 2'-SeMe-dG modified octamer DNA | | Descriptor: | DNA (5'-D(*GP*TP*(XUG)P*TP*AP*CP*AP*C)-3') | | Authors: | Sheng, J, Gan, J, Salon, J, Huang, Z. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis and crystal structure of 2'-se-modified guanosine containing DNA.
J.Org.Chem., 75, 2010
|
|
4HNJ
 
 | |
2AMI
 
 | | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | | Descriptor: | Caltractin | | Authors: | Hu, H.T, Fagan, P.A, Bunick, C.G, Sheehan, J.H, Chazin, W.J. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function.
J.Biol.Chem., 281, 2006
|
|
5WPQ
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5WPT
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed II conformation at 3.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5WPV
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs at 3.59 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
6BCJ
 
 | | cryo-EM structure of TRPM4 in apo state with short coiled coil at 3.1 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCL
 
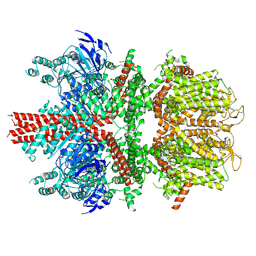 | | cryo-EM structure of TRPM4 in apo state with long coiled coil at 3.5 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCQ
 
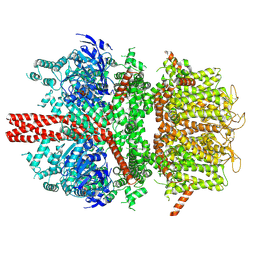 | | cryo-EM structure of TRPM4 in ATP bound state with long coiled coil at 3.3 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCO
 
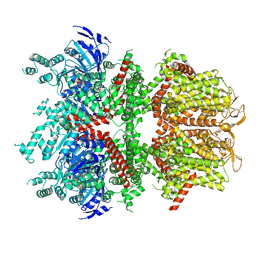 | | cryo-EM structure of TRPM4 in ATP bound state with short coiled coil at 2.9 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
8HVT
 
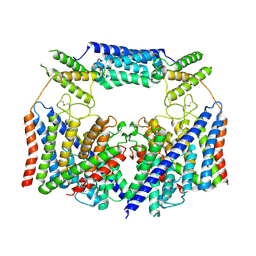 | | Structure of the human CLC-7/Ostm1 complex reveals a novel state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1, ... | | Authors: | Zhang, Z.X, Chen, L, He, J, She, J. | | Deposit date: | 2022-12-27 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human CLC-7/Ostm1 complex reveals a novel state
To Be Published
|
|
4EBZ
 
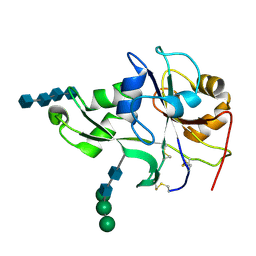 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBY
 
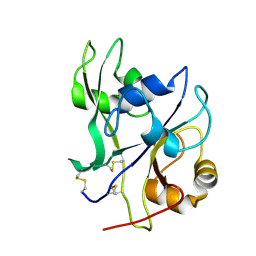 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
6Z18
 
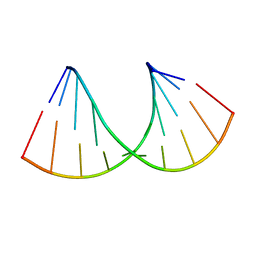 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | Descriptor: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | Authors: | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
1MZH
 
 | | QR15, an Aldolase | | Descriptor: | Deoxyribose-phosphate aldolase, PHOSPHATE ION | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aquifex Aeolicus Aldolase,
Northeast Structural Genomics Consortium Target
QR15
To be Published
|
|
1VCR
 
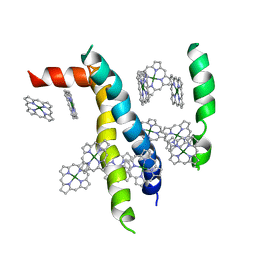 | | An icosahedral assembly of light-harvesting chlorophyll a/b protein complex from pea thylakoid membranes | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein AB80 | | Authors: | Hino, T, Kanamori, E, Shen, J.-R, Kouyama, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | An icosahedral assembly of the light-harvesting chlorophyll a/b protein complex from pea chloroplast thylakoid membranes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8EFO
 
 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFB
 
 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
6KMW
 
 | | Structure of PSI from H. hongdechloris grown under white light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
7Y3F
 
 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2022-06-10 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
7D32
 
 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D33
 
 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
