7DHG
 
 | | Crystal structure of SARS-CoV-2 Orf9b complex with human TOM70 | | Descriptor: | Mitochondrial import receptor subunit TOM70, ORF9b protein | | Authors: | Gao, X, Zhu, K, Qin, B, Olieric, V, Wang, M, Cui, S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions.
Nat Commun, 12, 2021
|
|
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | Descriptor: | ACETATE ION, Protein 2C | | Authors: | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2021-02-24 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
1DD1
 
 | |
1ZOQ
 
 | | IRF3-CBP complex | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Qin, B, Lin, K. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of IRF-3 in complex with CBP.
Structure, 13, 2005
|
|
9JE9
 
 | | Crystal structure of a amidase that can hydrolase PU plastic | | Descriptor: | Amidase | | Authors: | Li, Z.S, Fang, S.T, Zheng, Z.R, Xia, W, Zhou, H.Y, Li, W.S, You, S, Han, X, Liu, W.D. | | Deposit date: | 2024-09-02 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Engineering of a Urethanase for Enhanced Depolymerization of Polyurethane
Acs Catalysis, 15, 2025
|
|
3W95
 
 | |
5KWB
 
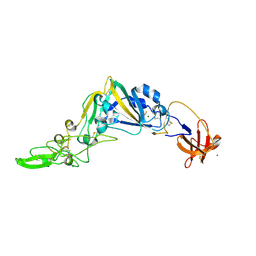 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
8X77
 
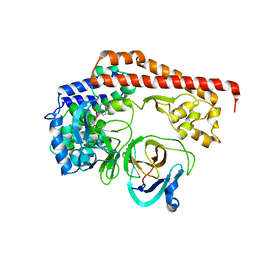 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
8X8Q
 
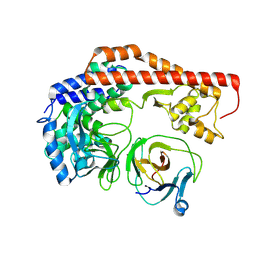 | | Structure of enterovirus protease in complex host factor | | Descriptor: | 2A protein (Fragment), Actin-histidine N-methyltransferase, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-28 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
7F60
 
 | |
7F90
 
 | |
7FEX
 
 | |
3X0F
 
 | |
3X0E
 
 | |
3X0G
 
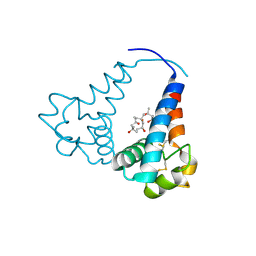 | |
6M40
 
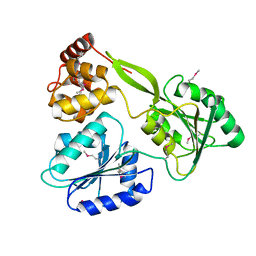 | | Crystal structure of the NS3-like helicase from Alongshan virus | | Descriptor: | NS3-like protein | | Authors: | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2020-03-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
5XGQ
 
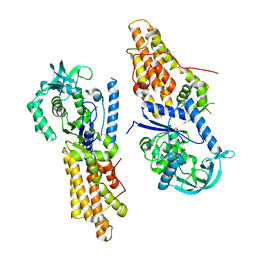 | |
5H7Y
 
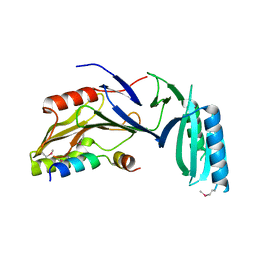 | |
3VGX
 
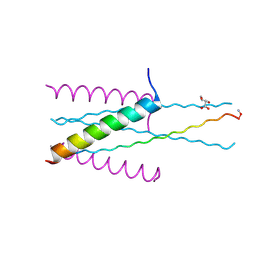 | | Structure of gp41 T21/Cp621-652 | | Descriptor: | ACETIC ACID, Envelope glycoprotein gp160, GLYCEROL | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of critical residues for viral entry and inhibition through structural Insight of HIV-1 fusion inhibitor CP621-652.
J.Biol.Chem., 287, 2012
|
|
3VH7
 
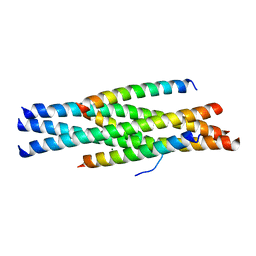 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P21 | | Descriptor: | CP32M, Envelope glycoprotein gp160, MAGNESIUM ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3STQ
 
 | |
3VWB
 
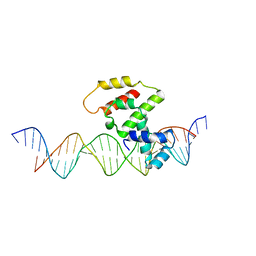 | | Crystal structure of VirB core domain (Se-Met derivative) complexed with the cis-acting site (5-BRU modifications) upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator virB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
5H7Z
 
 | |
3VGY
 
 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P321 | | Descriptor: | CP32M, Envelope glycoprotein gp160, SULFATE ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3W3C
 
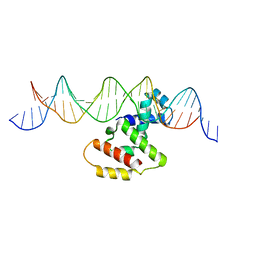 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
