8K0F
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0E
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
3TJT
 
 | | Crystal Structure Analysis of the superoxide dismutase from Clostridium difficile | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Lei, C, Ying, T.L, Wang, H.F, Tan, X.S. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of the superoxide dismutase from Clostridium difficile
To be Published
|
|
8KC9
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to cyclosporin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, DSN-MLE-MLE-MVA-BMT-ABA-SAR-MLE-VAL-MLE-ALA, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
4N0G
 
 | | Crystal Structure of PYL13-PP2CA complex | | Descriptor: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | Authors: | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
4JYY
 
 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | Descriptor: | AZIDE ION, FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
4JZG
 
 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
4JZ2
 
 | | Crystal structure of Co ion substituted SOD2 from Clostridium difficile | | Descriptor: | COBALT (II) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Co ion substituted SOD2 from Clostridium difficile
To be Published
|
|
5Y6R
 
 | | Crystal structure of CSFV NS5B | | Descriptor: | GLYCEROL, Genome polyprotein, SULFATE ION | | Authors: | Li, W, Wu, B. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Classical Swine Fever Virus NS5B Reveals a Novel N-Terminal Domain
J. Virol., 92, 2018
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
8AP0
 
 | | ForT Mutant T138V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 4-azanyl-1~{H}-pyrazole-3,5-dicarboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | Authors: | Li, W, Naismith, J.H. | | Deposit date: | 2022-08-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
8AOZ
 
 | | ForT Mutant L24A | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-3-[(2~{S},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]-1~{H}-pyrazole-5-carboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | Authors: | Li, W, Naismith, J.H. | | Deposit date: | 2022-08-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
6JCI
 
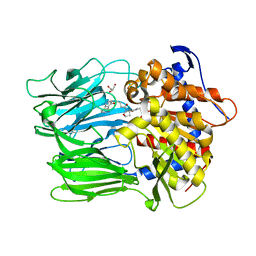 | |
7CY6
 
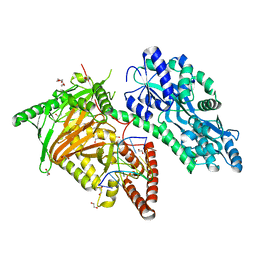 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY7
 
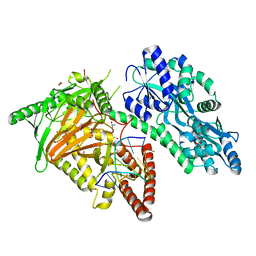 | | Crystal Structure of CMD1 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY5
 
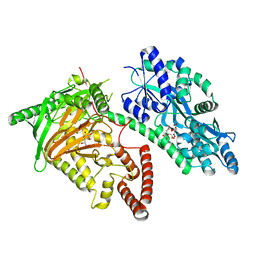 | | Crystal Structure of CMD1 in complex with vitamin C | | Descriptor: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY8
 
 | | Crystal Structure of CMD1 in complex with 5mC-DNA and vitamin C | | Descriptor: | 1,2-ETHANEDIOL, ASCORBIC ACID, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY4
 
 | | Crystal Structure of CMD1 in apo form | | Descriptor: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
4U86
 
 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U84
 
 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
3EP2
 
 | | Model of Phe-tRNA(Phe) in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
3UDC
 
 | | Crystal structure of a membrane protein | | Descriptor: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | Authors: | Li, W, Ge, J, Yang, M. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.355 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
