2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX0
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX4
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX3
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
7X9D
 
 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
3NPS
 
 | |
3SO3
 
 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
2L65
 
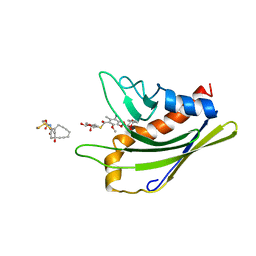 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
2EVN
 
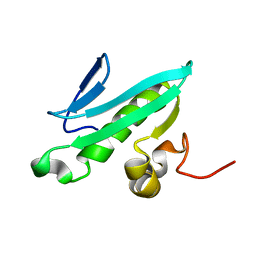 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
4ANW
 
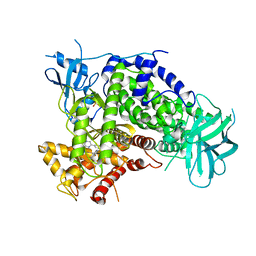 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-6-{4-CHLORO-3-[(2,3-DIFLUOROPHENYL)SULFAMOYL]PHENYL}-N-METHYLPYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4ANV
 
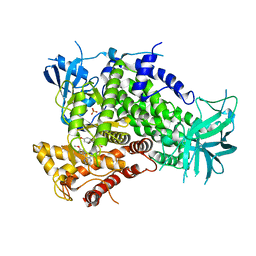 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 2-{4-[(4'-METHOXYBIPHENYL-3-YL)SULFONYL]PIPERAZIN-1-YL}-3-(4-METHOXYPHENYL)PYRAZINE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4ANX
 
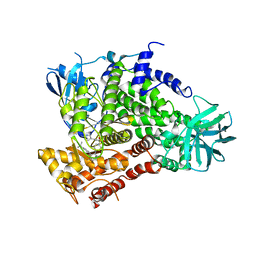 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 5-{3-[(4-{3-[4-(1-methylethyl)phenyl]pyrazin-2-yl}piperazin-1-yl)sulfonyl]phenyl}pyrimidin-2-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4BBF
 
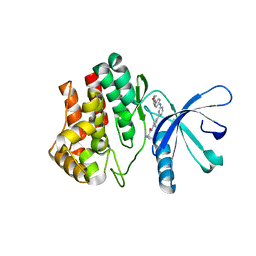 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | (2R)-N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]pyrrolidine-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ANU
 
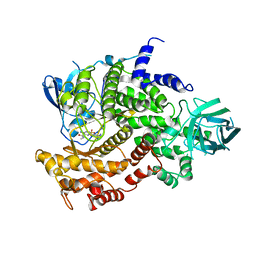 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-N-METHYL-6-[3-(1H-TETRAZOL-5-YL)PHENYL]PYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4BBE
 
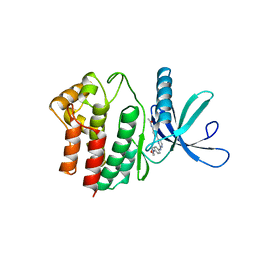 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]ethanamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6CBJ
 
 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
6CBP
 
 | |
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
6VTU
 
 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBI
 
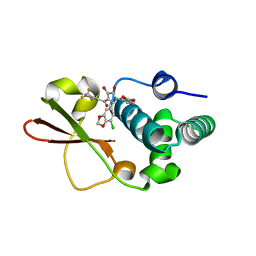 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
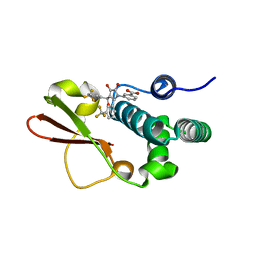 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
