3Q30
 
 | | Human Squalene synthase in complex with (2R,3R)-2-Carboxymethoxy-3-[5-(2-naphthalenyl)pentyl]aminocarbonyl-3-[5-(2-naphthalenyl)pentyloxy]propionic acid | | Descriptor: | (2R,3R)-2-(carboxymethoxy)-4-{[5-(naphthalen-2-yl)pentyl]amino}-3-{[5-(naphthalen-2-yl)pentyl]oxy}-4-oxobutanoic acid, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
3Q2Z
 
 | | Human Squalene synthase in complex with N-[(3R,5S)-7-Chloro-5-(2,3-dimethoxyphenyl)-1-neopentyl-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepine-3-acetyl]-L-aspartic acid | | Descriptor: | N-{[(3R,5S)-7-chloro-5-(2,3-dimethoxyphenyl)-1-(2,2-dimethylpropyl)-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepin-3-yl]acetyl}-L-aspartic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
4GOH
 
 | |
4GPF
 
 | |
4GPH
 
 | |
4GPC
 
 | |
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | Descriptor: | MYRISTIC ACID, Protein A46 | | Authors: | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | Deposit date: | 2015-11-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
3I9T
 
 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
5G0J
 
 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | Descriptor: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0G
 
 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | Descriptor: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0F
 
 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | Descriptor: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0H
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | Descriptor: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0I
 
 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker cleaved) in complex with Nexturastat A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HDAC6, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
3I9U
 
 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithioerythritol (DTE) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
3I8R
 
 | | Crystal structure of the heme oxygenase from Corynebacterium diphtheriae (HmuO) in complex with heme binding ditiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-10 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
9CEY
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | Descriptor: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF3
 
 | | Parasitella parasitica Fanzor (PpFz) State 4 | | Descriptor: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | Descriptor: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein,Spizellomyces punctatus Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
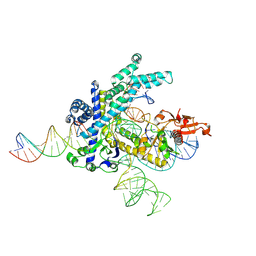 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CER
 
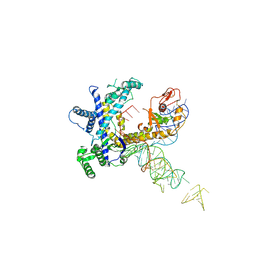 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
2ZNS
 
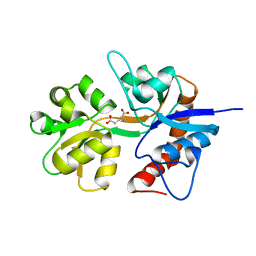 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3QXM
 
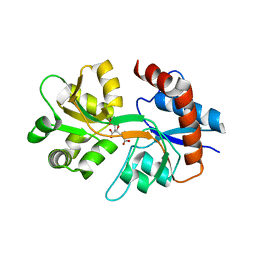 | | Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVG
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVK
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FUZ
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with L-glutamate in space group P1 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
