7SD0
 
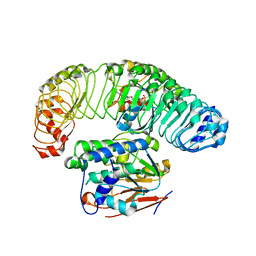 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
2M1B
 
 | |
4KC3
 
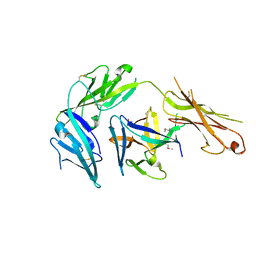 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LMY
 
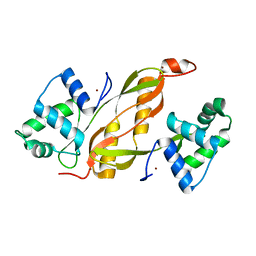 | | Structure of GAS PerR-Zn-Zn | | Descriptor: | Peroxide stress regulator PerR, FUR family, ZINC ION | | Authors: | Lin, C.S, Chao, S.Y, Nix, J.C, Tseng, H.L, Tsou, C.C, Fei, C.H, Ciou, H.S, Jeng, U.S, Lin, Y.S, Chuang, W.J, Wu, J.J, Wang, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct structural features of the peroxide response regulator from group a streptococcus drive DNA binding
Plos One, 9, 2014
|
|
6PAS
 
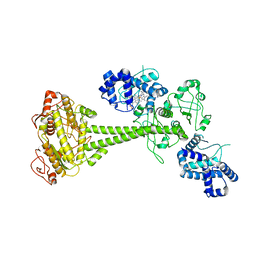 | | Inactive State of Manduca sexta soluble guanylate cyclase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble guanylyl cyclase alpha-1 subunit, Soluble guanylyl cyclase beta-1 subunit | | Authors: | Yokom, A.L, Horst, B.G, Marletta, M.A, Hurley, J.H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife, 8, 2019
|
|
6PAT
 
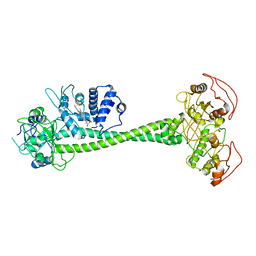 | | Active State of Manduca sexta soluble Guanylate Cyclase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble guanylyl cyclase alpha-1 subunit, Soluble guanylyl cyclase beta-1 subunit | | Authors: | Yokom, A.L, Horst, B.G, Marletta, M.A, Hurley, J.H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife, 8, 2019
|
|
3CRW
 
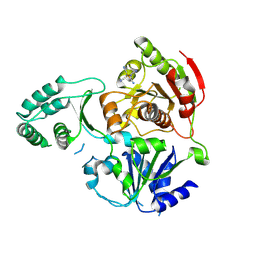 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3CRV
 
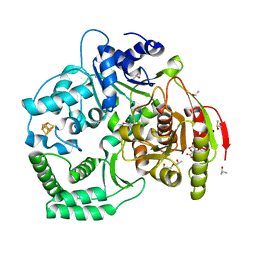 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3DPL
 
 | |
3HQL
 
 | |
3HVE
 
 | |
3HTM
 
 | |
3HQH
 
 | |
3HQI
 
 | |
3HSV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATHx-MacroH2ASBCpep2 | | Descriptor: | Core histone macro-H2A.1, SULFATE ION, Speckle-type POZ protein, ... | | Authors: | Zhuang, M, Schulman, B.A, Miller, D. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases.
Mol.Cell, 36, 2009
|
|
3HQM
 
 | |
3HU6
 
 | |
3IVQ
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
3QKS
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3PXJ
 
 | | Tandem Ig repeats of Dlar | | Descriptor: | Tyrosine-protein phosphatase Lar | | Authors: | Biersmith, B.H, Bouyain, S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3003 Å) | | Cite: | The Immunoglobulin-like Domains 1 and 2 of the Protein Tyrosine Phosphatase LAR Adopt an Unusual Horseshoe-like Conformation.
J.Mol.Biol., 408, 2011
|
|
3PXH
 
 | | Tandem Ig domains of tyrosine phosphatase LAR | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Biersmith, B.H, Bouyain, S. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0009 Å) | | Cite: | The Immunoglobulin-like Domains 1 and 2 of the Protein Tyrosine Phosphatase LAR Adopt an Unusual Horseshoe-like Conformation.
J.Mol.Biol., 408, 2011
|
|
3QKU
 
 | | Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKT
 
 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
