6PLK
 
 | |
7N1H
 
 | | CryoEM structure of Venezuelan equine encephalitis virus VLP in complex with the LDLRAD3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
7N1I
 
 | | CryoEM structure of Venezuelan equine encephalitis virus VLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, E1 envelope glycoprotein, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
6C74
 
 | |
6C6Q
 
 | |
4OII
 
 | |
4PDC
 
 | |
4OIE
 
 | |
4OIG
 
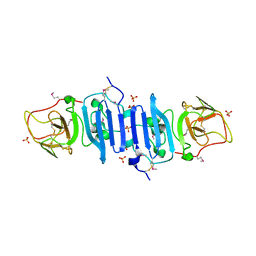 | |
2HG0
 
 | | Structure of the West Nile Virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
2FWO
 
 | |
2GRK
 
 | |
7K9K
 
 | |
7K9H
 
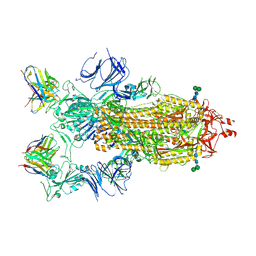 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7K9I
 
 | |
7K9J
 
 | |
7KYL
 
 | | Powassan virus Envelope protein DIII in complex with neutralizing Fab POWV-80 | | Descriptor: | CHLORIDE ION, Envelope protein domain III, POWV-80 Fab heavy chain, ... | | Authors: | Errico, J.M, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses.
J.Exp.Med., 218, 2021
|
|
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
3IRC
 
 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
3I50
 
 | | Crystal structure of the West Nile Virus envelope glycoprotein in complex with the E53 antibody Fab | | Descriptor: | Envelope glycoprotein, murine heavy chain (IgG3) of E53 monoclonal antibody Fab, murine kappa light chain of E53 monoclonal antibody Fab | | Authors: | Nybakken, G.E, Warren, J.T, Chen, B.R, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2009-07-03 | | Release date: | 2009-10-27 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody.
Embo J., 28, 2009
|
|
3HXO
 
 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | Descriptor: | Aptamer ARC1172, von Willebrand factor | | Authors: | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | Deposit date: | 2009-06-21 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
3IXX
 
 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
3HXQ
 
 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | Descriptor: | Aptamer ARC1172, von Willebrand Factor | | Authors: | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | Deposit date: | 2009-06-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
7JX6
 
 | |
4FFZ
 
 | | Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). | | Descriptor: | DENV1-E111 fab fragment (heavy chain), DENV1-E111 fab fragment (light chain), Envelope protein E | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
