7ASX
 
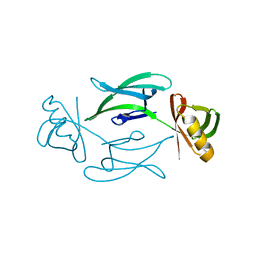 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chip 1) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
7ASI
 
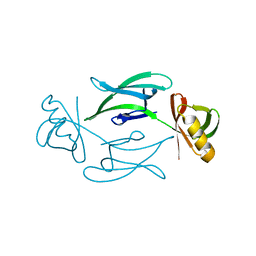 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chips 1+2) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
9G25
 
 | | snR30 snoRNP - State 1 - Utp23-Krr1-deltaC3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14-A, H/ACA ribonucleoprotein complex subunit CBF5, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
9G28
 
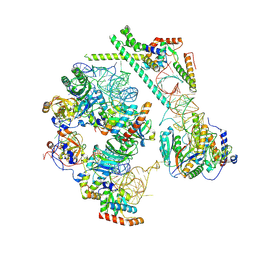 | | snR30 snoRNP - State 2 - Utp23-Krr1-deltaC3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14-A, H/ACA ribonucleoprotein complex subunit CBF5, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
9G33
 
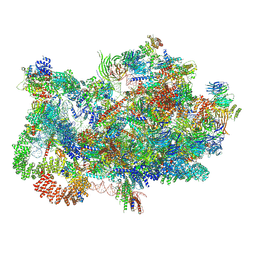 | | Stalled 90S - Utp23-Krr1-deltaC3 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-11 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
1URC
 
 | | Cyclin A binding groove inhibitor Ace-Arg-Lys-Leu-Phe-Gly | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, PEPTIDE INHIBITOR | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-10-28 | | Release date: | 2003-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, biological activity and structural analysis of cyclic peptide inhibitors targeting the substrate recruitment site of cyclin-dependent kinase complexes.
Org. Biomol. Chem., 2, 2004
|
|
6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
6PQ7
 
 | |
6OD9
 
 | | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CESIUM ION, MAGNESIUM ION, ... | | Authors: | Jones, C.P, Tran, B, Ferre-D'Amare, A.R. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.102 Å) | | Cite: | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4G5Z
 
 | |
4G6M
 
 | |
4G6K
 
 | |
4G6J
 
 | |
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV8
 
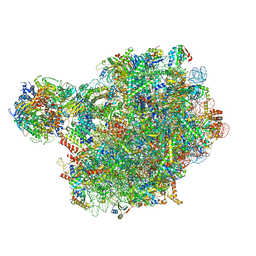 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV2
 
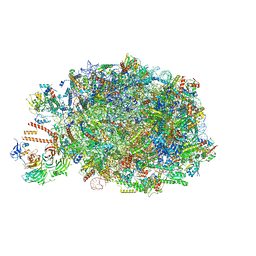 | | Chaetomium thermophilum pre-60S State 10 - pre-5S rotation with Ytm1-Erb1 | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV4
 
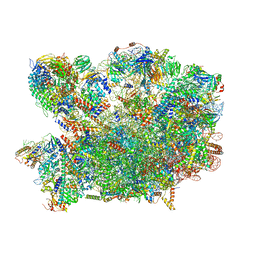 | | Chaetomium thermophilum pre-60S State 2 - pre-5S rotation with Rix1 complex - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV6
 
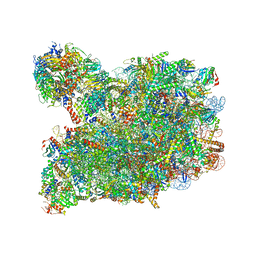 | | Chaetomium thermophilum pre-60S State 3 - post-5S rotation with Rix1 complex with Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV1
 
 | | Chaetomium thermophilum pre-60S State 6 - pre-5S rotation - L1 intermediate - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV5
 
 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV3
 
 | | Chaetomium thermophilum pre-60S State 9 - pre-5S rotation - immature H68/H69 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PTW
 
 | | Chaetomium thermophilum Rix1-complex | | Descriptor: | Pre-rRNA-processing protein IPI3, Pre-rRNA-processing protein RIX1 | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PUW
 
 | | Chaetomium thermophilum Las1-Grc3-complex | | Descriptor: | Las1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV7
 
 | | Chaetomium thermophilum pre-60S State 1 - pre-5S rotation (Arx1/Nog2 state) - Composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
