6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
3BS8
 
 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
5CVC
 
 | | Structure of maize serine racemase | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | Authors: | Song, Y, Zou, L, Fan, J. | | Deposit date: | 2015-07-26 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of maize serine racemase with pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5H3D
 
 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
3VT1
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactose | | Descriptor: | Ricin B lectin, beta-D-galactopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VT2
 
 | | Crystal structure of Ct1,3Gal43A in complex with isopropy-beta-D-thiogalactoside | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VSF
 
 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | Descriptor: | GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VSZ
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
4GQ2
 
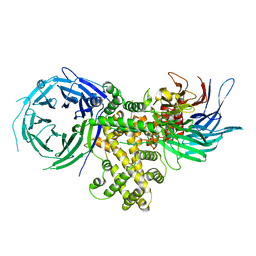 | | S. pombe Nup120-Nup37 complex | | Descriptor: | Nucleoporin nup120, Nup37 | | Authors: | Liu, X, Mitchell, J, Wozniak, R, Blobel, G, Fan, J. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evolution of the membrane-coating module of the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GQ1
 
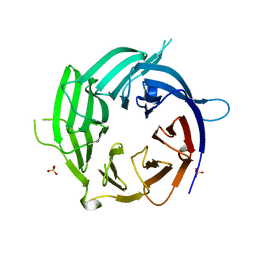 | | Nup37 of S. pombe | | Descriptor: | Nup37, SULFATE ION | | Authors: | Liu, X, Mitchell, J, Wozniak, R, Blobel, G, Fan, J. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evolution of the membrane-coating module of the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GNX
 
 | |
4GS4
 
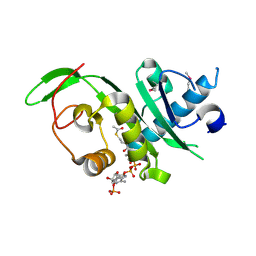 | | Structure of the alpha-tubulin acetyltransferase, alpha-TAT1 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase | | Authors: | Friedmann, D.R, Fan, J, Marmorstein, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Structure of the alpha-tubulin acetyltransferase, alpha TAT1, and implications for tubulin-specific acetylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OR6
 
 | |
4OR4
 
 | |
7E2V
 
 | | Crystal structure of MaDA-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA-3 | | Authors: | Gao, L, Du, X, Fan, J, Lei, X. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Enzymatic control of endo- and exo-stereoselective Diels-Alder reactions with broad substrate scope.
Nat Catal, 2021
|
|
8CD9
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 6 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family), SODIUM ION, ... | | Authors: | Rubesova, P, Brynda, J, Fanfrlik, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
5IYC
 
 | | Human core-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IYB
 
 | | Human core-PIC in the open state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IYD
 
 | | Human core-PIC in the initial transcribing state (no IIS) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
3NLT
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'S,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}- N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({(3S,4S)-4-[2-({2,2-difluoro-2-[(2R)-piperidin-2-yl]ethyl}amino)ethoxy]pyrrolidin-3-yl}methyl)-4-methylpyridin-2-amine, CACODYLIC ACID, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLU
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2,2-difluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
4RCG
 
 | | Crystal Structure Analysis of MTB PEPCK without Mn+2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brynda, J, Dostal, J, Snasel, J, Fanfrlik, J, Pichova, I, Machova, I. | | Deposit date: | 2014-09-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Studies of Phosphoenolpyruvate Carboxykinase from Mycobacterium tuberculosis.
Plos One, 10, 2015
|
|
4R43
 
 | | Crystal Structure Analysis of MTB PEPCK | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Brynda, J, Dostal, J, Pichova, I, Snasel, J, Fanfrlik, J, Machova, I. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Phosphoenolpyruvate Carboxykinase from Mycobacterium tuberculosis.
Plos One, 10, 2015
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
