4MRY
 
 | | Crystal Structure of Ca(2+)- discharged Y138F obelin mutant from Obelia longissima at 1.30 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MRX
 
 | | Crystal Structure of Y138F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PW8
 
 | | Human tryptophan 2,3-dioxygenase | | Descriptor: | COBALT (II) ION, Tryptophan 2,3-dioxygenase | | Authors: | Meng, B, Wu, D, Gu, J.H, Ouyang, S.Y, Ding, W, Liu, Z.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and functional analyses of human tryptophan 2,3-dioxygenase
Proteins, 82, 2014
|
|
1Z09
 
 | | Solution structure of km23 | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Ilangovan, U, Ding, W, Mulder, K, Hinck, A.P, Zuniga, J, Trbovich, J.A, Demeler, B, Groppe, J.C. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Homodimeric Dynein Light Chain km23.
J.Mol.Biol., 352 (2), 2005
|
|
1S1J
 
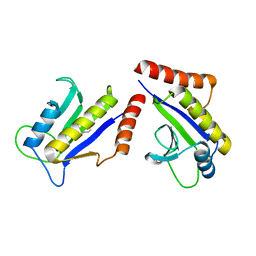 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S1S
 
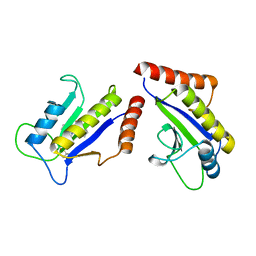 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | Descriptor: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | Authors: | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
7CYC
 
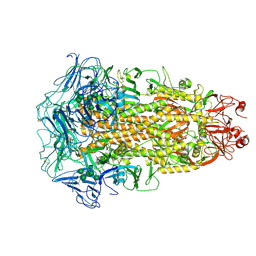 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
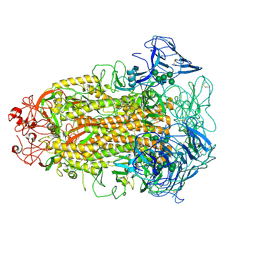 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
1IL1
 
 | | Crystal structure of G3-519, an anti-HIV monoclonal antibody | | Descriptor: | monoclonal antibody G3-519 (heavy chain), monoclonal antibody G3-519 (light chain) | | Authors: | Berry, M.B, Johnson, K.A, Radding, W, Fung, M, Liou, R, Phillips Jr, G.N. | | Deposit date: | 2001-05-07 | | Release date: | 2001-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an anti-HIV monoclonal Fab antibody fragment specific to a gp120 C-4 region peptide.
Proteins, 45, 2001
|
|
1J52
 
 | |
6IHB
 
 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
6IH9
 
 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
7XP7
 
 | | Crystal Structure of the Flavoprotein ColB1 Catalyzing Assembly Line-Tethered Cysteine Dehydrogenation | | Descriptor: | Cyclohexanecarboxyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, X.Y, Tang, Z.J, Liu, W, Ma, M. | | Deposit date: | 2022-05-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanistic Insights into ColB1, a Flavoprotein Functioning in-trans in the 2,2'-Bipyridine Assembly Line for Cysteine Dehydrogenation.
Acs Chem.Biol., 18, 2023
|
|
8IL0
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 | | Descriptor: | Glycosyltransferase | | Authors: | Dai, Y, Li, P, Qiao, H, Xia, M, Liu, W, Fang, P. | | Deposit date: | 2023-03-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
6IM4
 
 | |
8J67
 
 | | Crystal structure of Toxoplasma gondii M2AP | | Descriptor: | MIC2-associated protein | | Authors: | Wang, F.F, Zhang, D.J, Zhang, S, Springer, T.A, Song, G.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
8J64
 
 | | Crystal structure of Toxoplasma gondii MIC2-M2AP complex | | Descriptor: | MIC2-associated protein, Micronemal protein MIC2 | | Authors: | Zhang, S, Wang, F.F, Zhang, D.J, Song, G.J, Springer, T.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
9UT8
 
 | | The full-length human sweet taste receptor TAS1R2 and TAS1R3 in the apo state | | Descriptor: | Taste receptor type 1 member 2,Engineered red fluorescent protein mScarlet3, Taste receptor type 1 member 3,mNeonGreen | | Authors: | Shi, Z.J, Xu, W.X, Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-05-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural and functional characterization of human sweet taste receptor.
Nature, 2025
|
|
9UT9
 
 | | The VFT domains of human sweet taste receptor TAS1R2 and TAS1R3 in the apo state | | Descriptor: | Taste receptor type 1 member 2,Engineered red fluorescent protein mScarlet3, Taste receptor type 1 member 3,mNeonGreen | | Authors: | Shi, Z.J, Xu, W.X, Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-05-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural and functional characterization of human sweet taste receptor.
Nature, 2025
|
|
9UTC
 
 | | The VFT domains of human sweet taste receptor TAS1R2 and TAS1R3 in the sucralose-bound state | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Taste receptor type 1 member 2,Taste receptor type 1 member 2,Engineered red fluorescent protein mScarlet3, Taste receptor type 1 member 3,mNeonGreen | | Authors: | Shi, Z.J, Xu, W.X, Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-05-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural and functional characterization of human sweet taste receptor.
Nature, 2025
|
|
9UTA
 
 | | The transmembrane domains of human sweet taste receptor TAS1R2 and TAS1R3 in the apo state | | Descriptor: | Taste receptor type 1 member 2,Engineered red fluorescent protein mScarlet3, Taste receptor type 1 member 3,mNeonGreen | | Authors: | Shi, Z.J, Xu, W.X, Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-05-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural and functional characterization of human sweet taste receptor.
Nature, 2025
|
|
9UTB
 
 | | The full-length human sweet taste receptor TAS1R2 and TAS1R3 in the sucralose-bound state | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Taste receptor type 1 member 2,Taste receptor type 1 member 2,Taste receptor type 1 member 2,Engineered red fluorescent protein mScarlet3, Taste receptor type 1 member 3,mNeonGreen | | Authors: | Shi, Z.J, Xu, W.X, Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-05-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural and functional characterization of human sweet taste receptor.
Nature, 2025
|
|
9L1N
 
 | | Structure of Western equine encephalitis virus 71V1658 strain VLP in complex with human PCDH10 EC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid glycoprotein, E1 glycoprotein, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | Deposit date: | 2024-12-15 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
6XCJ
 
 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
