7K4D
 
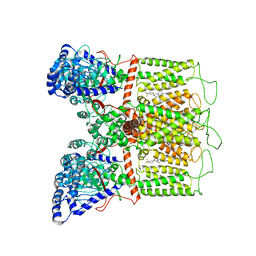 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 3OG | | Descriptor: | 5-[(4-{trans-4-hydroxy-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4B
 
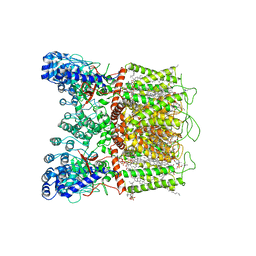 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor cis-22a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[cis-4-(3-methylphenyl)cyclohexyl]-4-(pyridin-3-yl)piperazine, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4A
 
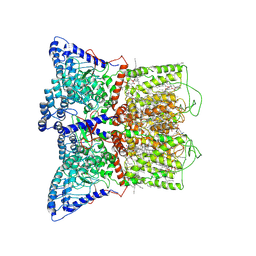 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
3NM7
 
 | | Crystal Structure of Borrelia burgdorferi Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Graebsch, A, Roche, S, Kostrewa, D, Niessing, D. | | Deposit date: | 2010-06-22 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Of Bits and Bugs - on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure.
Plos One, 5, 2010
|
|
1QXZ
 
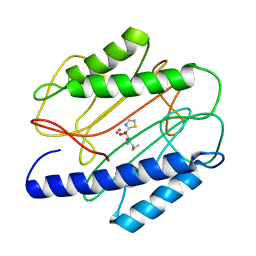 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle inhibitor 119 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-(1,3-THIAZOL-2-YL)BUTANE-1,1-DIOL, COBALT (II) ION, methionyl aminopeptidase | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QXY
 
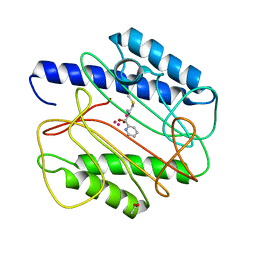 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle 618 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-PYRIDIN-2-YLBUTANE-1,1-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
7D2K
 
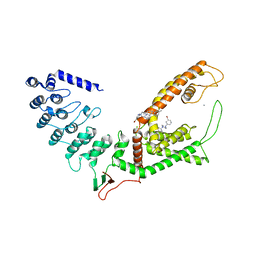 | | Crystal structure of rat TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromanylpyridin-3-yl)-4-[4-(3-methylphenyl)cyclohexyl]piperazin-4-ium, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
1QXW
 
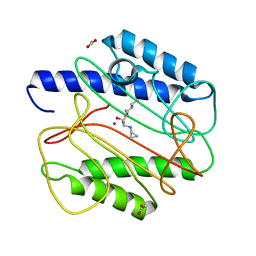 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1P83
 
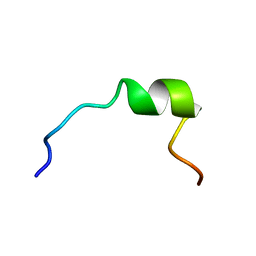 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
1P82
 
 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
