4B16
 
 | |
5DIY
 
 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
5DJS
 
 | | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M | | Descriptor: | Tetratricopeptide TPR_2 repeat protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
6X83
 
 | |
6X82
 
 | | Crystal Structure of TNFalpha with isoquinoline compound 4 | | Descriptor: | 8-[4-(2-{5-[(4-methylpiperazin-1-yl)methyl]-2-(1H-pyrrolo[3,2-c]pyridin-3-yl)phenoxy}ethyl)phenyl]isoquinoline, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
6X86
 
 | | Crystal Structure of TNFalpha with indolinone compound 11 | | Descriptor: | 3-[(6-{2-[(3R)-4-(hydroxyacetyl)-3-methylpiperazin-1-yl]pyrimidin-5-yl}-2,2-dimethyl-3-oxo-2,3-dihydro-1H-indol-1-yl)methyl]pyridine-2-carbonitrile, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
6X81
 
 | |
2GA2
 
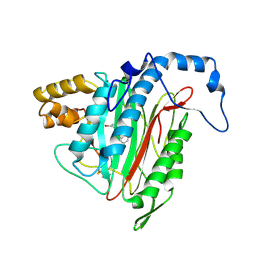 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4V97
 
 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2012-04-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V8J
 
 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8FT8
 
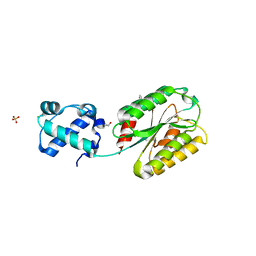 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, SUMO tag-free preparation | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FT6
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, SUMO tag-free preparation. | | Descriptor: | CITRIC ACID, IODIDE ION, SULFATE ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8VE9
 
 | | IsPETase - ACCCETN mutant - CombiPETase | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8VEM
 
 | | IsPETase - ACCE mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8VEK
 
 | | IsPETase - ACC mutant | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8VEL
 
 | | IsPETase - ACCCC mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8CRU
 
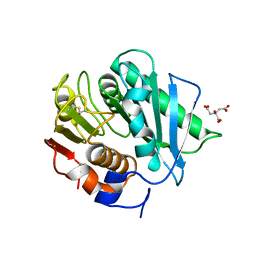 | | PETase Ancestral Sequence Reconstruction 008 | | Descriptor: | CITRIC ACID, Poly(ethylene terephthalate) hydrolase | | Authors: | Joho, Y, Royan, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ancestral Sequence Reconstruction Identifies Structural Changes Underlying the Evolution of Ideonella sakaiensis PETase and Variants with Improved Stability and Activity.
Biochemistry, 62, 2023
|
|
6X85
 
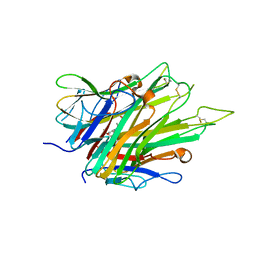 | | Crystal Structure of TNFalpha with indolinone compound 9 | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2,2-dimethyl-1,2-dihydro-3H-indol-3-one, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
8FZ4
 
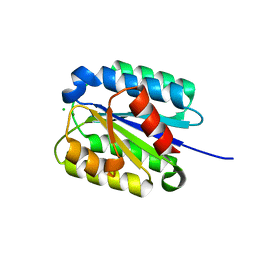 | | The von Willebrand factor A domain of Anthrax toxin receptor 2 | | Descriptor: | Anthrax toxin receptor 2, CHLORIDE ION, MAGNESIUM ION | | Authors: | Pedroza-Romo, M.J, Soleimani, S, Lewis, A, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8QBQ
 
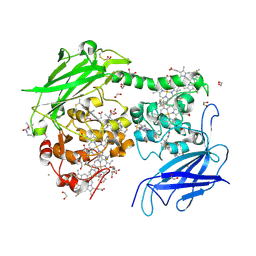 | | Crystal structure of the outer membrane decaheme cytochrome MtrC (A430Boc-Lys) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Nash, B.W, Lockwood, C.J, Whiting, K, Butt, J.N, Clarke, T.A, Edwards, M.J. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Genetic Code Expansion in Shewanella oneidensis MR-1 Allows Site-Specific Incorporation of Bioorthogonal Functional Groups into a c -Type Cytochrome.
Acs Synth Biol, 2024
|
|
8QBZ
 
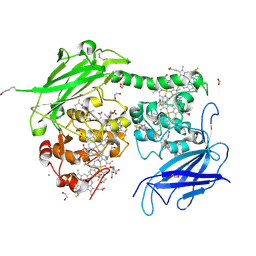 | | Crystal structure of the outer membrane decaheme cytochrome MtrC (E344Boc-Lys) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Edwards, M.J, Lockwood, C.J, Whiting, K, Nash, B.W, Butt, J.N, Clarke, T.A. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Genetic Code Expansion in Shewanella oneidensis MR-1 Allows Site-Specific Incorporation of Bioorthogonal Functional Groups into a c -Type Cytochrome.
Acs Synth Biol, 2024
|
|
8QC9
 
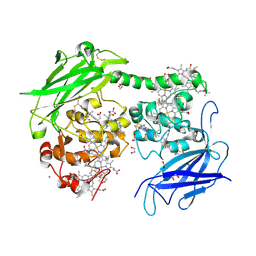 | | Crystal structure of the outer membrane decaheme cytochrome MtrC (A293Boc-Lys) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Nash, B.W, Lockwood, C.J, Edwards, M.J, Whiting, K, Butt, J.N, Clarke, T.A. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetic Code Expansion in Shewanella oneidensis MR-1 Allows Site-Specific Incorporation of Bioorthogonal Functional Groups into a c -Type Cytochrome.
Acs Synth Biol, 2024
|
|
6BY0
 
 | |
9FGP
 
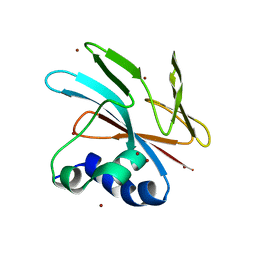 | |
2YGY
 
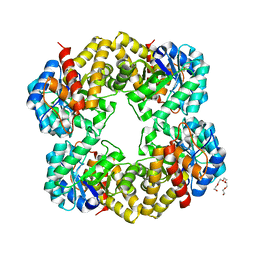 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
