6G0C
 
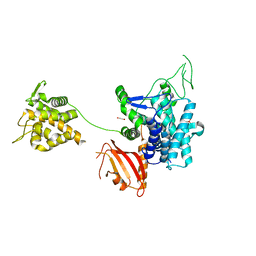 | | Crystal structure of SdeA catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kalayil, S, Bhogaraju, S, Basquin, J, Dikic, I. | | Deposit date: | 2018-03-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature, 557, 2018
|
|
7NY7
 
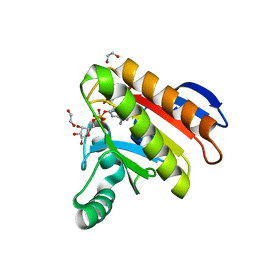 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NY6
 
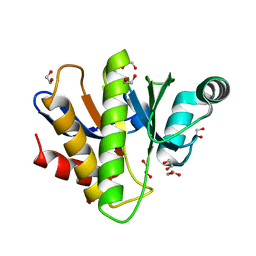 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Histone macroH2A1.1 | | Authors: | Knobloch, G, Guberovic, I, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5MZN
 
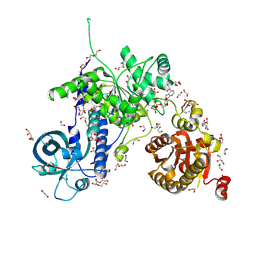 | | Helicase Sen1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leonaite, B, Basquin, J, Conti, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family.
EMBO J., 36, 2017
|
|
5NNZ
 
 | | Crystal structure of human ODA16 | | Descriptor: | Dynein assembly factor with WDR repeat domains 1 | | Authors: | Lorentzen, E, Taschner, T, Basquin, J. | | Deposit date: | 2017-04-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Purification and crystal structure of human ODA16: Implications for ciliary import of outer dynein arms by the intraflagellar transport machinery.
Protein Sci., 29, 2020
|
|
5MZH
 
 | | Crystal structure of ODA16 from Chlamydomonas reinhardtii | | Descriptor: | Dynein assembly factor with WDR repeat domains 1, SULFATE ION | | Authors: | Lorentzen, E, Taschner, M, Basquin, J, Mourao, A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46.
J. Biol. Chem., 292, 2017
|
|
6IAN
 
 | | T. brucei IFT22/74/81 GTP-bound crystal structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 74, Intraflagellar transport protein 81, ... | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
6IA7
 
 | | T. brucei IFT22 GTP-bound crystal structure | | Descriptor: | CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, ... | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
6IAE
 
 | | T. brucei IFT22 GDP-bound crystal structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, MAGNESIUM ION | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
4JA0
 
 | | Crystal structure of the invertebrate bi-functional purine biosynthesis enzyme PAICS at 2.8 A resolution | | Descriptor: | Phosphoribosylaminoimidazole carboxylase, SULFATE ION | | Authors: | Taschner, M, Basquin, J, Benda, C, Lorentzen, E. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the invertebrate bifunctional purine biosynthesis enzyme PAICS at 2.8 angstrom resolution.
Proteins, 81, 2013
|
|
2JA9
 
 | | Structure of the N-terminal deletion of yeast exosome component Rrp40 | | Descriptor: | EXOSOME COMPLEX EXONUCLEASE RRP40 | | Authors: | Oddone, A, Lorentzen, E, Basquin, J, Gasch, A, Rybin, V, Conti, E, Sattler, M. | | Deposit date: | 2006-11-24 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Characterization of the Yeast Exosome Component Rrp40
Embo Rep., 8, 2007
|
|
2HSN
 
 | | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes | | Descriptor: | GU4 nucleic-binding protein 1, Methionyl-tRNA synthetase, cytoplasmic | | Authors: | Simader, H, Koehler, C, Basquin, J, Suck, D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.
Nucleic Acids Res., 34, 2006
|
|
5NNV
 
 | |
5NMO
 
 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
2WCW
 
 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | ACETATE ION, AMMONIUM ION, HJC | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
2VNU
 
 | | Crystal structure of Sc Rrp44 | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP)-3', EXOSOME COMPLEX EXONUCLEASE RRP44, MAGNESIUM ION, ... | | Authors: | Lorentzen, E, Basquin, J, Conti, E. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the Rnase II Nuclease Family
Mol.Cell, 29, 2008
|
|
2WCZ
 
 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | CHLORIDE ION, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
2WIZ
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WJ0
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, COBALT HEXAMMINE(III), HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WIW
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | 5'-D(*DC*DG*DG*DA*DT*DA*DT*DC*DC*DGP)-3', GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
7ZEG
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7GMP | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
7ZEE
 
 | | Human cytosolic 5' nucleotidase IIIB | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methylguanosine phosphate-specific 5'-nucleotidase, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
7ZEH
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7Guanine | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, D-ribulose, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
6H25
 
 | | Human nuclear RNA exosome EXO-10-MPP6 complex | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Gerlach, P, Schuller, J.M, Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Distinct and evolutionary conserved structural features of the human nuclear exosome complex.
Elife, 7, 2018
|
|
6H4G
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei: E311A, T318R, V319A mutant bound to cAMP in the A site | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-07-21 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.143517 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
