3VR5
 
 | | Crystal structure of nucleotide-free Enterococcus hirae V1-ATPase [eV1(L)] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B, V-type sodium ATPase subunit D, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
2D6P
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with T-antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6O
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6N
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6K
 
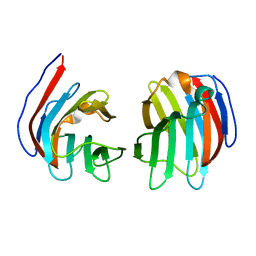 | | Crystal structure of mouse galectin-9 N-terminal CRD (crystal form 1) | | Descriptor: | lectin, galactose binding, soluble 9 | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2ZHL
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 2) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHK
 
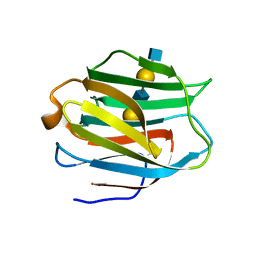 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 1) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHM
 
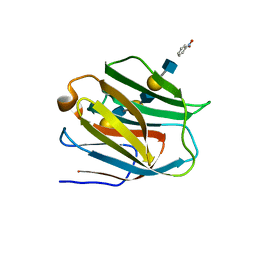 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
7C95
 
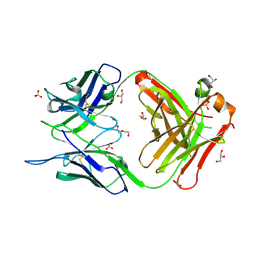 | | Crystal structure of the anti-human podoplanin antibody Fab fragment | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Nakamura, S, Suzuki, K, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
2ZHN
 
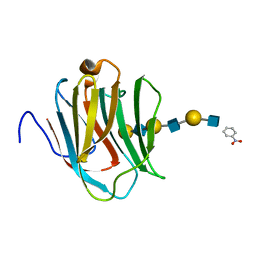 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 2) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, GALECTIN-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
7C94
 
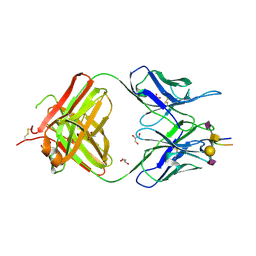 | | Crystal structure of the anti-human podoplanin antibody Fab fragment complex with glycopeptide | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Suzuki, K, Nakamura, S, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
3AON
 
 | | Crystal structure of the central axis (NtpD-NtpG) in the catalytic portion of Enterococcus hirae V-type sodium ATPase | | Descriptor: | NITRATE ION, V-type sodium ATPase subunit D, V-type sodium ATPase subunit G | | Authors: | Saijo, S, Arai, S, Hossain, K.M.M, Yamato, I, Kakinuma, Y, Ishizuka-Katsura, Y, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-04 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the central axis DF complex of the prokaryotic V-ATPase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | Descriptor: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | Authors: | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7DQC
 
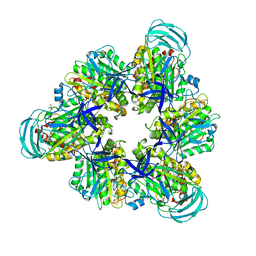 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
8GW8
 
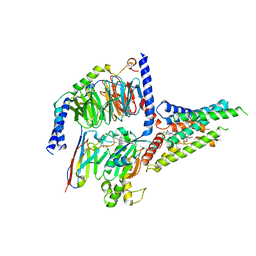 | | the human PTH1 receptor bound to an intracellular biased agonist | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Okamoto, H.H, Nureki, O. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Class B1 GPCR activation by an intracellular agonist.
Nature, 618, 2023
|
|
4YB9
 
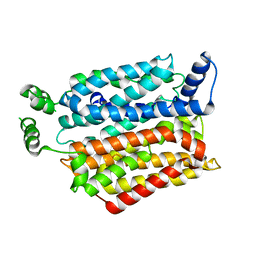 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
4YBQ
 
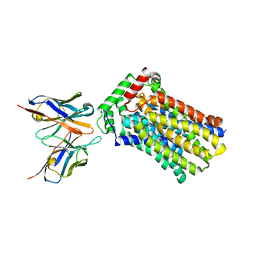 | | Rat GLUT5 with Fv in the outward-open form | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5, antibody Fv fragment heavy chain, ... | | Authors: | Nomura, N, Shimamura, T, Iwata, S. | | Deposit date: | 2015-02-19 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5
Nature, 526, 2015
|
|
3ORN
 
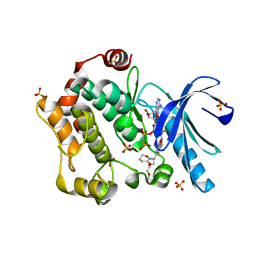 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4987655 and MgAMP-PNP | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(3-oxo-1,2-oxazinan-2-yl)methyl]benzamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OS3
 
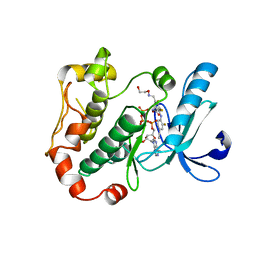 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4858061 and MgATP | | Descriptor: | 2-[(4-ethynyl-2-fluorophenyl)amino]-3,4-difluoro-N-(2-hydroxyethoxy)-5-{[(2-hydroxyethoxy)imino]methyl}benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7XMB
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
9JB2
 
 | |
9JAZ
 
 | |
9JB0
 
 | |
9JB1
 
 | |
