2UVS
 
 | | High Resolution Solid-state NMR structure of Kaliotoxin | | Descriptor: | POTASSIUM CHANNEL TOXIN ALPHA-KTX 3.1 | | Authors: | Korukottu, J, Lange, A, Vijayan, V, Schneider, R, Pongs, O, Becker, S, Baldus, M, Zweckstetter, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Conformational Plasticity in Ion Channel Recognition of a Peptide Toxin
To be Published
|
|
8B2U
 
 | | Crystal structure of SUDV VP40 in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Matrix protein VP40 | | Authors: | Werner, A.-D, Krapoth, N, Norris, M.J, Heine, A, Klebe, G, Ollmann Saphire, E, Becker, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a Crystallographic Screening to Identify Sudan Virus VP40 Ligands.
Acs Omega, 9, 2024
|
|
8AYT
 
 | | Crystal structure of SUDV VP40 W95A mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Steinchen, W, Werel, L, Kowalski, K, Essen, L.-O, Becker, S. | | Deposit date: | 2022-09-03 | | Release date: | 2023-09-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SUDV VP40 W95A mutant
To Be Published
|
|
8AYU
 
 | | Crystal structure of SUDV VP40 L117A mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Steinchen, W, Werel, L, Kowalski, K, Essen, L.-O, Becker, S. | | Deposit date: | 2022-09-03 | | Release date: | 2023-09-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of SUDV VP40 L117A mutant
To Be Published
|
|
8B1S
 
 | | co-crystal of SUDV VP40 with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Matrix protein VP40 | | Authors: | Werner, A.-D, Krapoth, N, Norris, M.J, Heine, A, Klebe, G, Ollmann Saphire, E, Becker, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a Crystallographic Screening to Identify Sudan Virus VP40 Ligands.
Acs Omega, 9, 2024
|
|
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
2GZU
 
 | |
1Y1U
 
 | | Structure of unphosphorylated STAT5a | | Descriptor: | Signal transducer and activator of transcription 5A | | Authors: | Neculai, D, Neculai, A.M, Verrier, S, Straub, K, Klumpp, K, Pfitzner, E, Becker, S. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure of the unphosphorylated STAT5a dimer
J.Biol.Chem., 280, 2005
|
|
1XA8
 
 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1X6M
 
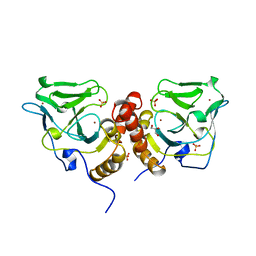 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1OJ5
 
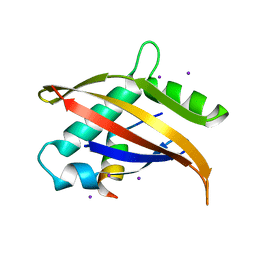 | | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | | Descriptor: | IODIDE ION, SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 6, STEROID RECEPTOR COACTIVATOR 1A | | Authors: | Razeto, A, Ramakrishnan, V, Giller, K, Lakomek, N, Carlomagno, T, Griesinger, C, Lodrini, M, Litterst, C.M, Pftizner, E, Becker, S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain
J.Mol.Biol., 336, 2004
|
|
2CM5
 
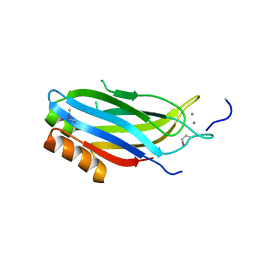 | | crystal structure of the C2B domain of rabphilin | | Descriptor: | CALCIUM ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
1URQ
 
 | | Crystal structure of neuronal Q-SNAREs in complex with R-SNARE motif of Tomosyn | | Descriptor: | M-TOMOSYN ISOFORM, SYNAPTOSOMAL-ASSOCIATED PROTEIN 25, SYNTAXIN 1A | | Authors: | Pobbati, A, Razeto, A, Becker, S, Fasshauer, D. | | Deposit date: | 2003-10-31 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibitory Role of Tomosyn in Exocytosis
J.Biol.Chem., 279, 2004
|
|
1XSW
 
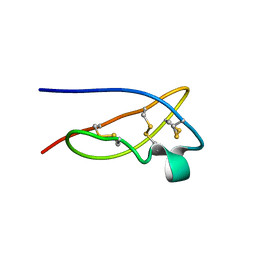 | | The solid-state NMR structure of Kaliotoxin | | Descriptor: | Kaliotoxin 1 | | Authors: | Lange, A, Becker, S, Seidel, K, Giller, K, Pongs, O, Baldus, M. | | Deposit date: | 2004-10-20 | | Release date: | 2005-04-05 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | A Concept for Rapid Protein-Structure Determination by Solid-State NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1GL2
 
 | | Crystal structure of an endosomal SNARE core complex | | Descriptor: | ENDOBREVIN, SYNTAXIN 7, SYNTAXIN 8, ... | | Authors: | Antonin, W, Becker, S, Jahn, R, Schneider, T.R. | | Deposit date: | 2001-08-22 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Endosomal Snare Complex Reveals Common Structural Principles of All Snares.
Nat.Struct.Biol., 9, 2001
|
|
3RIO
 
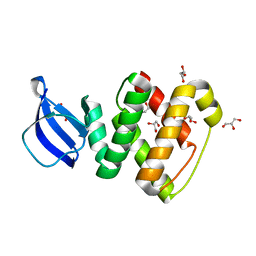 | | Crystal structure of GlcT CAT-PRDI | | Descriptor: | GLYCEROL, PtsGHI operon antiterminator | | Authors: | Himmel, S, Grosse, C, Wolff, S, Becker, S. | | Deposit date: | 2011-04-14 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the RBD-PRDI fragment of the antiterminator protein GlcT.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7ABT
 
 | |
2CA7
 
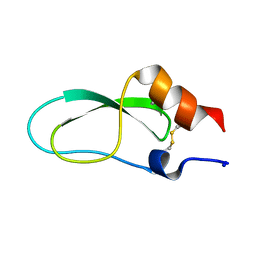 | | Conkunitzin-S1 Is The First Member Of A New Kunitz-Type Neurotoxin Family- Structural and Functional Characterization | | Descriptor: | CONKUNITZIN-S1 | | Authors: | Bayrhuber, M, Vijayan, V, Ferber, M, Graf, R, Korukottu, J, Imperial, J, Garrett, J.E, Olivera, B.M, Terlau, H, Zweckstetter, M, Becker, S. | | Deposit date: | 2005-12-19 | | Release date: | 2006-01-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization.
J. Biol. Chem., 280, 2005
|
|
2CHD
 
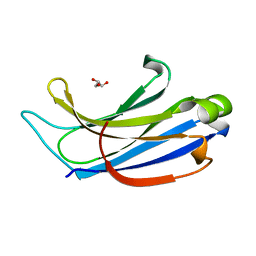 | | Crystal structure of the C2A domain of Rabphilin-3A | | Descriptor: | GLYCEROL, RABPHILIN-3A | | Authors: | Biadene, M, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the C2A Domain of Rabphilin-3A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CM6
 
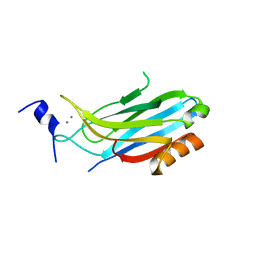 | | crystal structure of the C2B domain of rabphilin3A | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
4MIV
 
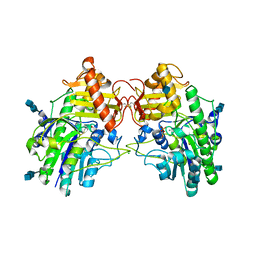 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MHX
 
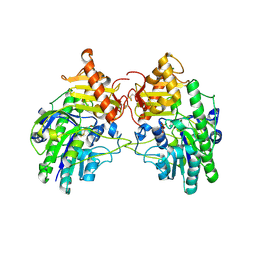 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8AEX
 
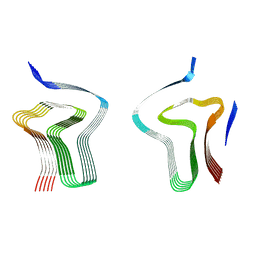 | | Lipidic alpha-synuclein fibril - polymorph L3A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADV
 
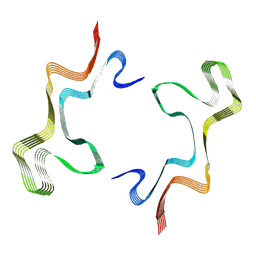 | | Lipidic alpha-synuclein fibril - polymorph L1B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADS
 
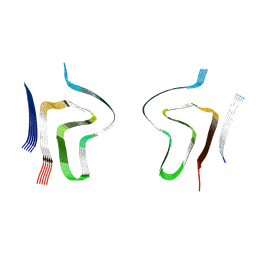 | | Lipidic alpha-synuclein fibril - polymorph L2B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
