2H9H
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
2HH5
 
 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7ETJ
 
 | | C5 portal vertex in the partially-enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ET2
 
 | | Human Cytomegalovirus, C12 portal | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETM
 
 | | C6 portal vertex in the enveloped virion capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETO
 
 | | C1 CVSC-binding penton vertex in the virion capsid of Human Cytomegalovirus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7KM5
 
 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
7ET3
 
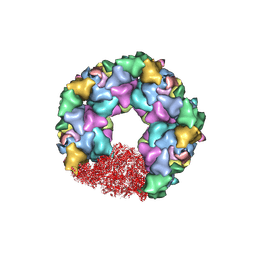 | | C5 portal vertex in the enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
2WV6
 
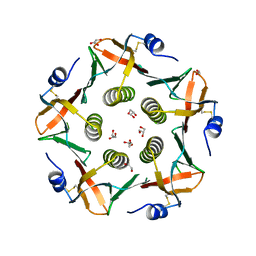 | | Crystal structure of the cholera toxin-like B-subunit from Citrobacter freundii to 1.9 angstrom | | Descriptor: | 1,2-ETHANEDIOL, CFXB, GLYCEROL | | Authors: | Jansson, L, Lebens, M, Imberty, A, Varrot, A, Teneberg, S. | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Carbohydrate Binding Specificities and Crystal Structure of the Cholera Toxin-Like B-Subunit from Citrobacter Freundii.
Biochimie, 92, 2010
|
|
3NLV
 
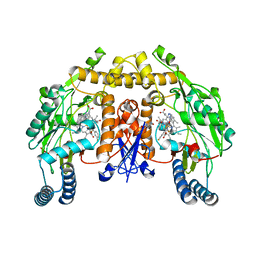 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-(2,2-Difluoro-2-(3-fluorophenyl)ethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-(2-{[2,2-difluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
1O20
 
 | |
5U1Q
 
 | | Grb7-SH2 with bicyclic peptide inhibitor | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 7, LYS-PHE-GLU-GLY-TYR-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
3B3P
 
 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
1O4S
 
 | |
1O4T
 
 | |
3NLW
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-(2,2-Difluoro-2-(piperidin-2-yl)ethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({(3S,4S)-4-[2-({2,2-difluoro-2-[(2R)-piperidin-2-yl]ethyl}amino)ethoxy]pyrrolidin-3-yl}methyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
5YSZ
 
 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
5U06
 
 | | Grb7-SH2 with bicyclic peptide inhibitor containing a pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, POTASSIUM ION, bicyclic peptide inhibitor: LYS-PHE-GLU-GLY-CMF-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, J.A. | | Deposit date: | 2016-11-22 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2H6M
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | N-ACETYL-LEUCYL-ALANYL-ALANYL-(N,N-DIMETHYL)-GLUTAMINE-(1,4-DIOXO-3,4-DIHYDRO-1H-PHTHALAZIN-2-YL)METHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Picornain 3C | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors.
J.Mol.Biol., 361, 2006
|
|
7E8N
 
 | | Crystal structure of Type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554 | | Descriptor: | CITRIC ACID, Citrate synthase | | Authors: | Park, S.-H, Lee, C.W, Bae, D.-W, Lee, J.H. | | Deposit date: | 2021-03-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Int.J.Biol.Macromol., 183, 2021
|
|
8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
