6I0U
 
 | | Crystal structure of DmTailor in complex with U6 RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*UP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
3ZN1
 
 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6Y02
 
 | | Thrombin in complex with 13k | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(5-bromanylfuran-2-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Thrombin in complex with 13k
To be published
|
|
3B4F
 
 | | Carbonic anhydrase inhibitors. Interaction of 2-(hydrazinocarbonyl)-3-phenyl-1H-indole-5-sulfonamide with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | 2-(hydrazinocarbonyl)-3-phenyl-1H-indole-5-sulfonamide, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Guzel, o, Temperini, c, Innocenti, a, Scozzafava, A, Salman, a, Supuran, c.t. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of 2-(hydrazinocarbonyl)-3-phenyl-1H-indole-5-sulfonamide with 12 mammalian isoforms: kinetic and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3AXK
 
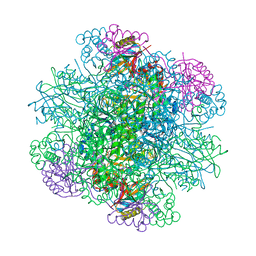 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
6G25
 
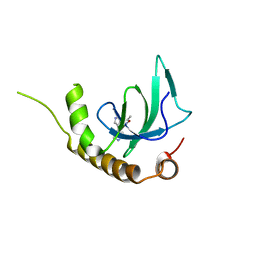 | | X-ray structure of NSD3-PWWP1 in complex with compound 4 | | Descriptor: | 3,5-dimethyl-4-(4-pyridin-4-yl-1~{H}-pyrazol-3-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6HJ6
 
 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6SPG
 
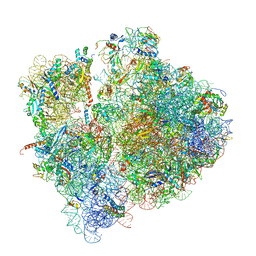 | | Pseudomonas aeruginosa 70s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6G2B
 
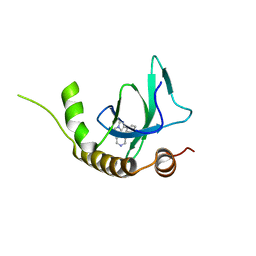 | | X-ray structure of NSD3-PWWP1 in complex with compound 8 | | Descriptor: | 4-(3-methyl-5-phenyl-imidazol-4-yl)pyridine, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6VX8
 
 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5YPY
 
 | | Crystal structure of IlvN. Val-1c | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-04 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
6W01
 
 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6UKC
 
 | |
6G2O
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound BI-9321 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6Y9H
 
 | | Thrombin in complex with D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2), DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-03-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2)
To be published
|
|
7TX5
 
 | |
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1RTA
 
 | |
3ZQJ
 
 | | Mycobacterium tuberculosis UvrA | | Descriptor: | UVRABC SYSTEM PROTEIN A, ZINC ION | | Authors: | Rossi, F, Khanduja, J.S, Bortoluzzi, A, Houghton, J, Sander, P, Guthlein, C, Davis, E.O, Springer, B, Bottger, E.C, Relini, A, Penco, A, Muniyappa, K, Rizzi, M. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Biological and Structural Characterization of Mycobacterium Tuberculosis Uvra Provides Novel Insights Into its Mechanism of Action
Nucleic Acids Res., 39, 2011
|
|
1RTB
 
 | |
7TX4
 
 | |
6YB6
 
 | | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c) | | Descriptor: | 2-(aminomethyl)-5-chloranyl-benzene-1,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c)
To be published
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
7KXL
 
 | | BTK1 SOAKED WITH COMPOUND 5, Y551 IS SEQUESTERED | | Descriptor: | 3-tert-butyl-N-({2-fluoro-4-[2-(1-methyl-1H-pyrazol-4-yl)-1H-imidazo[4,5-b]pyridin-7-yl]phenyl}methyl)-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
