8YZI
 
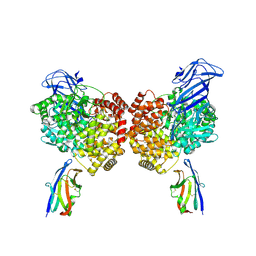 | | The structure of PDCoV RBD and dog APN complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Sun, J.Q, Niu, S. | | Deposit date: | 2024-04-07 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species.
Plos Pathog., 21, 2025
|
|
8YGG
 
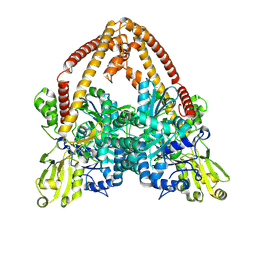 | | pP1192R-apo Closed state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGH
 
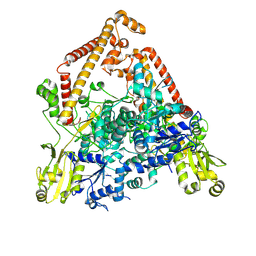 | | pP1192R-apo open state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGE
 
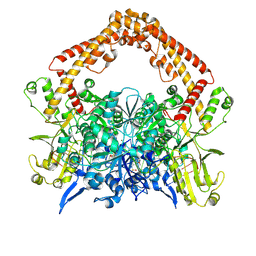 | | pP1192R-DNA-m-AMSA complex DNA binding/cleavage domain | | Descriptor: | DNA (12-mer), DNA (20-mer), DNA (8-mer), ... | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YIK
 
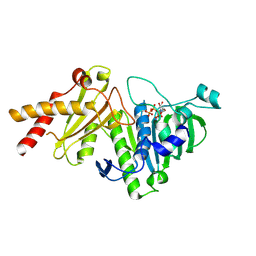 | | pP1192R-ATPase-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8XHE
 
 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XG1
 
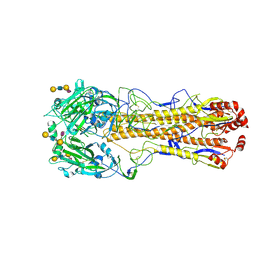 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XHG
 
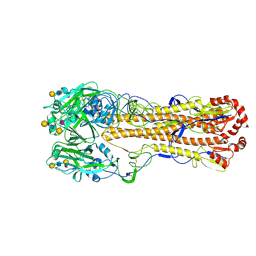 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XHH
 
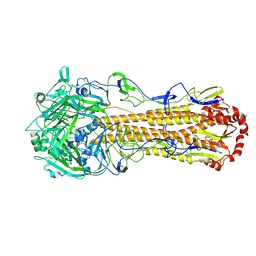 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7WBI
 
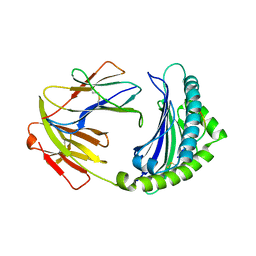 | | BF2*1901-FLU | | Descriptor: | Beta-2-microglobulin, ILE-ARG-HIS-GLU-ASN-ARG-MET-VAL-LEU, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7WBG
 
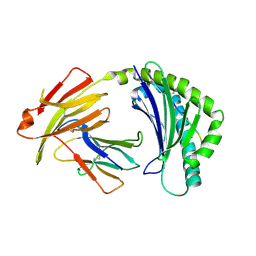 | | BF2*1901/RY8 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7W14
 
 | |
7W17
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234E) | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYK
 
 | |
7VYL
 
 | |
7VXH
 
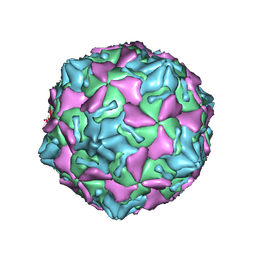 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYM
 
 | |
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VXZ
 
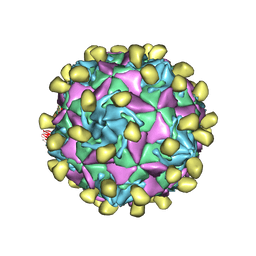 | |
7XXJ
 
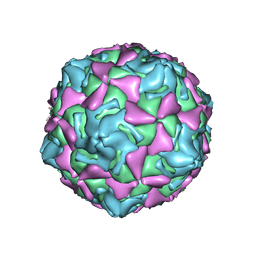 | | Echo 18 incubated with FcRn at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
