7QXW
 
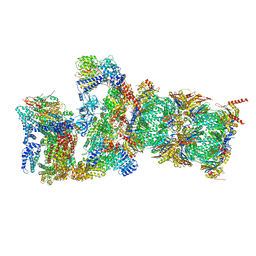 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
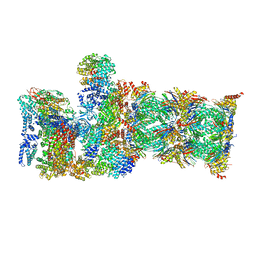 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
8ETR
 
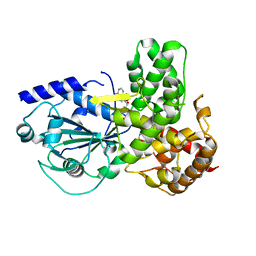 | | CryoEM Structure of NLRP3 NACHT domain in complex with G2394 | | Descriptor: | (6S,8R)-N-[(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)carbamoyl]-6-(methylamino)-6,7-dihydro-5H-pyrazolo[5,1-b][1,3]oxazine-3-sulfonamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Murray, J.M, Johnson, M.C. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Overcoming Preclinical Safety Obstacles to Discover ( S )- N -((1,2,3,5,6,7-Hexahydro- s -indacen-4-yl)carbamoyl)-6-(methylamino)-6,7-dihydro-5 H -pyrazolo[5,1- b ][1,3]oxazine-3-sulfonamide (GDC-2394): A Potent and Selective NLRP3 Inhibitor.
J.Med.Chem., 65, 2022
|
|
2P1W
 
 | |
2QKW
 
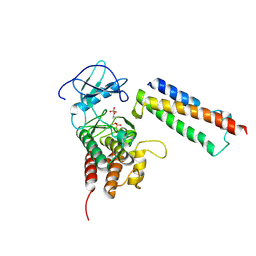 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | Descriptor: | Avirulence protein, Protein kinase | | Authors: | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
5UCW
 
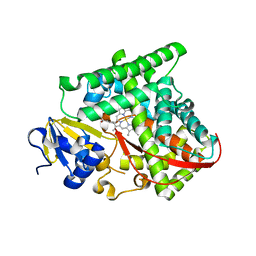 | |
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
6MKO
 
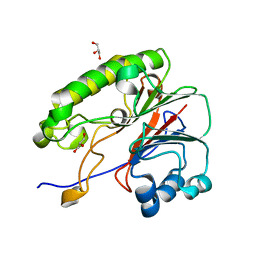 | |
6MKK
 
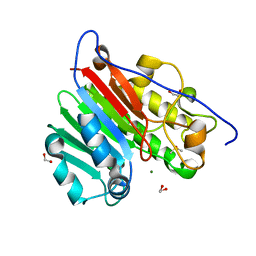 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
2Q8Y
 
 | |
6MKM
 
 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
7DZ9
 
 | | MbnABC complex | | Descriptor: | FE (III) ION, MbnA, MbnB, ... | | Authors: | Chao, D, Dan, Z, Yijun, G, Wei, C. | | Deposit date: | 2021-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
5Z5T
 
 | | The first bromodomain of BRD4 with compound BDF-2141 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5V
 
 | | The first bromodomain of BRD4 with compound BDF-1253 | | Descriptor: | Bromodomain-containing protein 4, N-{8-hydroxy-4-[(1H-imidazol-1-yl)methyl]quinolin-2-yl}acetamide | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5U
 
 | | The first bromodomain of BRD4 with compound BDF-2254 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinoline-6,8-diol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
7FC0
 
 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | Deposit date: | 2021-07-13 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | Descriptor: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
7VOA
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
4I0S
 
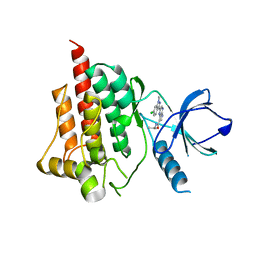 | |
4I0R
 
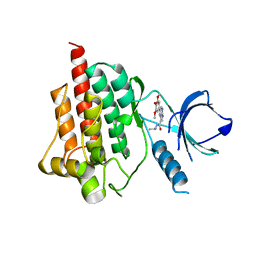 | | Crystal structure of spleen tyrosine kinase complexed with 2-(3,4,5-Trimethoxy-phenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid isopropylamide | | Descriptor: | N-(propan-2-yl)-2-(3,4,5-trimethoxyphenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Villasenor, A.G. | | Deposit date: | 2012-11-19 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
4IO0
 
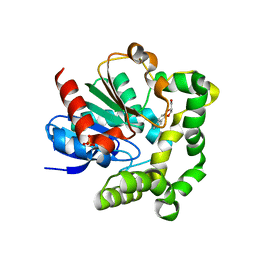 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | Descriptor: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
4O08
 
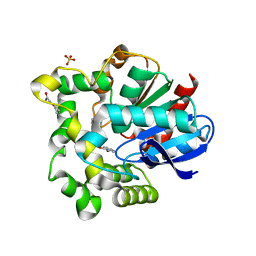 | |
