4BJB
 
 | | Crystal structure of human tankyrase 2 in complex with PJ-34 | | Descriptor: | GLYCEROL, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-04-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4BJC
 
 | | Crystal structure of human tankyrase 2 in complex with Rucaparib | | Descriptor: | Rucaparib, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-04-18 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
4AVW
 
 | | Crystal structure of human tankyrase 2 in complex with TIQ-A | | Descriptor: | 4H-thieno[2,3-c]isoquinolin-5-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
4B29
 
 | | Crystal structures of DMSP lyases RdDddP and RnDddQII | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DIMETHYLSULFONIOPROPIONATE LYASE, ... | | Authors: | Hehemann, J.H, Law, A, Redecke, L, Boraston, A.B. | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structures of Dmsp Lyases Rddddp and Rndddqii
To be Published
|
|
2JHI
 
 | | Structure of globular heads of M-ficolin complexed with N-acetyl-D- galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, FICOLIN-1 | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
4AT2
 
 | | The crystal structure of 3-ketosteroid-delta4-(5alpha)-dehydrogenase from Rhodococcus jostii RHA1 in complex with 4-androstene-3,17- dione | | Descriptor: | 3-KETOSTEROID-DELTA4-5ALPHA-DEHYDROGENASE, 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, ... | | Authors: | van Oosterwijk, N, Knol, J, Dijkhuizen, L, van der Geize, R, Dijkstra, B.W. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Catalytic Mechanism of 3-Ketosteroid-{Delta}4-(5Alpha)-Dehydrogenase from Rhodococcus Jostii Rha1 Genome.
J.Biol.Chem., 287, 2012
|
|
4AVU
 
 | | Crystal structure of human tankyrase 2 in complex with 6(5H)- phenanthridinone | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
4C1X
 
 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 6'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, SIALIDASE A | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Characterization of the Carbohydrate-Binding Module of Nana Sialidase, a Pneumococcal Virulence Factor.
Bmc Struct.Biol., 15, 2015
|
|
7YQT
 
 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR3
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQV
 
 | | pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
4BTL
 
 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
7YR1
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR2
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR0
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (interface) | | Descriptor: | Heavy chain of S309, IGK@ protein, Spike protein S1, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
4C34
 
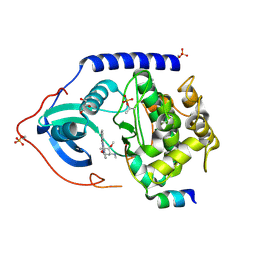 | | PKA-S6K1 Chimera with Staurosporine bound | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, GLYCEROL, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
7YQW
 
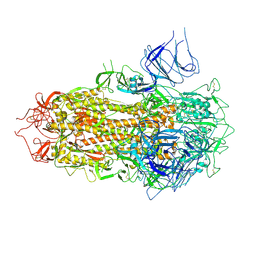 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
7YQX
 
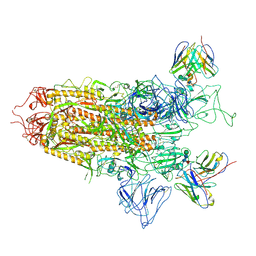 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
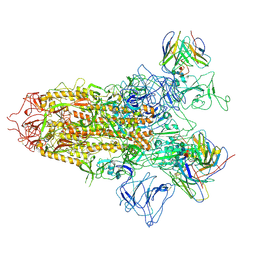 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
4BUS
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(4-oxo-3, 4-dihydroquinazolin-2-yl)phenoxy)acetic acid | | Descriptor: | 2-[4-(4-oxidanylidene-3H-quinazolin-2-yl)phenoxy]ethanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
4BUA
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(methylsulfanyl)phenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-(4-methylsulfanylphenyl)-3H-quinazolin-4-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
4BUW
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(2-oxo-1, 3-oxazolidin-3-yl)phenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-[4-(2-OXO-1,3-OXAZOLIDIN-3-YL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
