7GRX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-20 | | Descriptor: | 1-(2,4-difluorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRS
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-15 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRE
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-1 | | Descriptor: | 3C-like proteinase nsp5, 4-[3-(trifluoromethyl)-1H-pyrazol-5-yl]pyridine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A.M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRM
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-9 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRV
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-18 | | Descriptor: | (2S)-2-(2-fluorophenyl)-1,3-thiazolidin-4-one, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS1
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-24 | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS0
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | Descriptor: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | Descriptor: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4HQB
 
 | |
7JXX
 
 | | Structure of TTBK1 kinase domain in complex with Compound 3 | | Descriptor: | 4-(2-amino-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-11-yl)-2-methylbut-3-yn-2-ol, SODIUM ION, Tau-tubulin kinase 1 | | Authors: | Chodaprambil, J.V. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Tau Tubulin Kinase 1 (TTBK1) Inhibitors that Lower Tau Phosphorylation In Vivo.
J.Med.Chem., 64, 2021
|
|
7JXY
 
 | | Structure of TTBK1 kinase domain in complex with Compound 18 | | Descriptor: | (3S)-1-[1-(2-aminopyrimidin-4-yl)-1H-pyrazolo[4,3-c]pyridin-6-yl]-3-methylpent-1-yn-3-ol, Tau-tubulin kinase 1 | | Authors: | Chodaprambil, J.V. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Tau Tubulin Kinase 1 (TTBK1) Inhibitors that Lower Tau Phosphorylation In Vivo.
J.Med.Chem., 64, 2021
|
|
1OS2
 
 | | Ternary enzyme-product-inhibitor complexes of human MMP12 | | Descriptor: | ACETATE ION, ACETOHYDROXAMIC ACID, AZIDE ION, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Structures of Binary and Ternary Enzyme-Product-Inhibitor Complexes of Matrix Metalloproteinases
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
7JJC
 
 | |
7K61
 
 | | Cryo-EM structure of 197bp nucleosome aided by scFv | | Descriptor: | DNA (197-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.-R, Bai, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Mol.Cell, 81, 2021
|
|
4N5H
 
 | | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Esterase/lipase, ... | | Authors: | Bennett, M.D, Holland, R, Loo, T.S, Smith, C.A, Norris, G.E, Delabre, M.-L, Anderson, B.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001)
TO BE PUBLISHED
|
|
7K5Y
 
 | |
7K60
 
 | |
1QSO
 
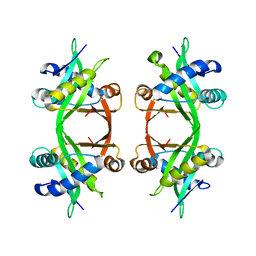 | | Histone Acetyltransferase HPA2 from Saccharomyces Cerevisiae | | Descriptor: | HPA2 HISTONE ACETYLTRANSFERASE | | Authors: | Angus-Hill, M.L, Dutnall, R.N, Tafrov, S.T, Sterngalnz, R, Ramakrishnan, V. | | Deposit date: | 1999-06-22 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily.
J.Mol.Biol., 294, 1999
|
|
7K5X
 
 | |
7KRN
 
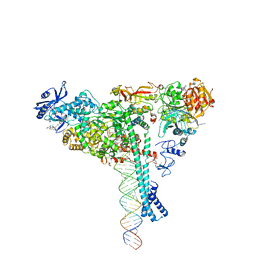 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K63
 
 | |
3U5V
 
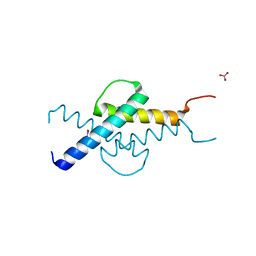 | | Crystal structure of Max-E47 | | Descriptor: | NITRATE ION, Protein max, Transcription factor E2-alpha chimera | | Authors: | Guarne, A, Ahmadpour, F, Gloyd, M. | | Deposit date: | 2011-10-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the minimalist max-e47 protein chimera.
Plos One, 7, 2012
|
|
7KRP
 
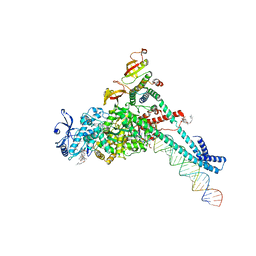 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRO
 
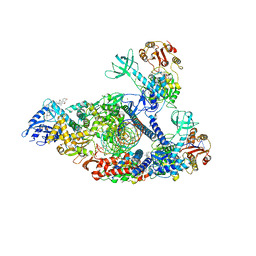 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1RMZ
 
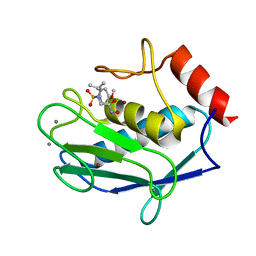 | | Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor NNGH at 1.3 A resolution | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational variability of matrix metalloproteinases: beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
