1M93
 
 | |
1OHH
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE complexed with the inhibitor protein IF1 | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Cabezon, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Bovine F1-ATPase in Complex with its Regulatory Protein If1
Nat.Struct.Biol., 10, 2003
|
|
1E1Q
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE AT 100K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-05-10 | | Release date: | 2000-06-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase Inhibited by Mg2+Adp and Aluminium Fluoride
Structure, 8, 2000
|
|
1E1R
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE INHIBITED BY MG2+ADP AND ALUMINIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-05-10 | | Release date: | 2000-06-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase Inhibited by Mg2+Adp and Aluminium Fluoride
Structure, 8, 2000
|
|
4UHR
 
 | | Thermostabilised HUMAN A2a Receptor with CGS21680 bound | | Descriptor: | 2-[P-(2-CARBOXYETHYL)PHENYLETHYL-AMINO]-5'-N-ETHYLCARBOXAMIDO ADENOSINE, THERMOSTABILISED HUMAN A2A RECEPTOR | | Authors: | Lebon, G, Edwards, P.C, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor.
Mol.Pharmacol., 87, 2015
|
|
4UG2
 
 | | Thermostabilised HUMAN A2a Receptor with CGS21680 bound | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[P-(2-CARBOXYETHYL)PHENYLETHYL-AMINO]-5'-N-ETHYLCARBOXAMIDO ADENOSINE, THERMOSTABILISED HUMAN A2A RECEPTOR | | Authors: | Lebon, G, Edwards, P.C, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-03-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor.
Mol.Pharmacol., 87, 2015
|
|
6IBL
 
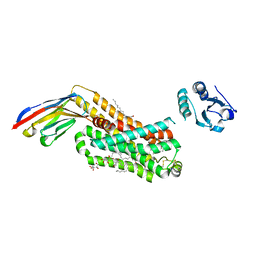 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
1H8E
 
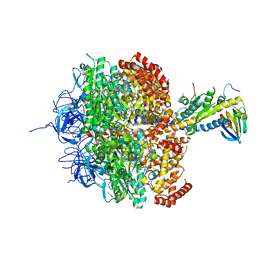 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
5G53
 
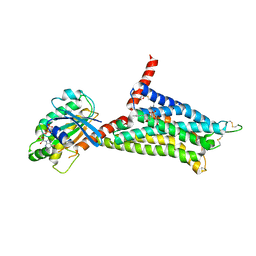 | | Structure of the adenosine A2A receptor bound to an engineered G protein | | Descriptor: | ADENOSINE RECEPTOR A2A, ENGINEERED DOMAIN OF HUMAN G ALPHA S LONG ISOFORM, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Carpenter, B, Nehme, R, Warne, T, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Adenosine A2A Receptor Bound to an Engineered G Protein
Nature, 536, 2016
|
|
6Q45
 
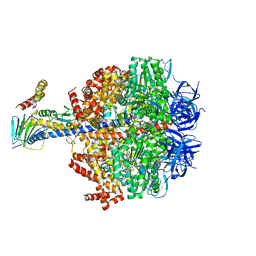 | | F1-ATPase from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Petri, J, Nakatani, Y, Montgomery, M.G, Ferguson, S.A, Aragao, D, Leslie, A.G.W, Heikal, A, Walker, J.E, Cook, G.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of F1-ATPase from the obligate anaerobe Fusobacterium nucleatum.
Open Biology, 9, 2019
|
|
1QO1
 
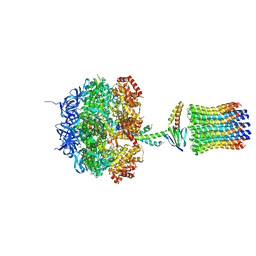 | | Molecular Architecture of the Rotary Motor in ATP Synthase from Yeast Mitochondria | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Stock, D, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1999-11-01 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture of the Rotary Motor in ATP Synthase
Science, 286, 1999
|
|
1E79
 
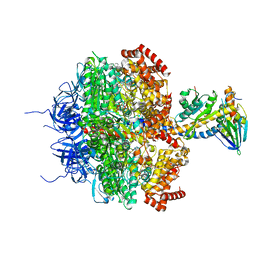 | | Bovine F1-ATPase inhibited by DCCD (dicyclohexylcarbodiimide) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, ... | | Authors: | Gibbons, C, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-08-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Central Stalk in Bovine F(1)-ATPase at 2.4 A Resolution.
Nat.Struct.Biol., 7, 2000
|
|
6ZMV
 
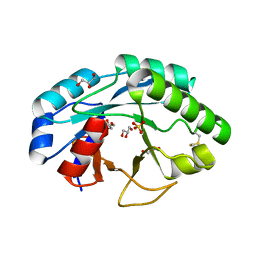 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
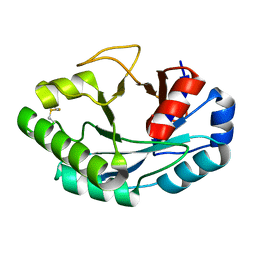 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
2CLY
 
 | | Subcomplex of the stator of bovine mitochondrial ATP synthase | | Descriptor: | ATP SYNTHASE B CHAIN, MITOCHONDRIAL, ATP SYNTHASE COUPLING FACTOR 6, ... | | Authors: | Kane Dickson, V, Silvester, J.A, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the Structure of the Stator of the Mitochondrial ATP Synthase.
Embo J., 25, 2006
|
|
8B2G
 
 | | SH3-like domain from Penicillium virgatum muramidase | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2F
 
 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | Descriptor: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2H
 
 | | Muramidase from Thermothielavioides terrestris, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2E
 
 | | Muramidase from Kionochaeta sp natural catalytic core | | Descriptor: | CADMIUM ION, Muramidase | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6F5D
 
 | | Trypanosoma brucei F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma subunit, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Gahura, O, Leslie, A.G.W, Zikova, A, Walker, J.E. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ATP synthase fromTrypanosoma bruceihas an elaborated canonical F1-domain and conventional catalytic sites.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8B2S
 
 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | Descriptor: | GH24 family muramidase, POTASSIUM ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6FOC
 
 | | F1-ATPase from Mycobacterium smegmatis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, T, Montgomery, M.G, Leslie, A.G.W, Cook, G.M, Walker, J.E. | | Deposit date: | 2018-02-06 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structure of the catalytic domain of the ATP synthase fromMycobacterium smegmatisis a target for developing antitubercular drugs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1N11
 
 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
1CIA
 
 | |
6GK0
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
