3HKO
 
 | | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40 | | Descriptor: | Calcium/calmodulin-dependent protein kinase with a kinase domain and 2 calmodulin-like EF hands, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Wasney, G, Vedadi, M, MacKenzie, F, Kozieradzki, I, Cossar, D, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-25 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40
To be Published
|
|
2YBD
 
 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
2YKM
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with a Difluoromethylbenzoxazole (DFMB) Pyrimidine Thioether derivative, a non-nucleoside RT inhibitor (NNRTI) | | Descriptor: | 2-[DIFLUORO-[(4-METHYL-PYRIMIDINYL)-THIO]METHYL]-BENZOXAZOLE, CALCIUM ION, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Boyer, J, Arnoult, E, Medebielle, M, Guillemont, J, Unge, T, Unge, J, Jochmans, D. | | Deposit date: | 2011-05-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Difluoromethylbenzoxazole Pyrimidine Thioether Derivatives: A Novel Class of Potent Non-Nucleoside HIV-1 Reverse Transcriptase Inhibitors.
J.Med.Chem., 54, 2011
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
2Y69
 
 | | Bovine heart cytochrome c oxidase re-refined with molecular oxygen | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, CHOLIC ACID, ... | | Authors: | Kaila, V.R.I, Oksanen, E, Goldman, A, Verkhovsky, M.I, Sundholm, D, Wikstrom, M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Combined Quantum Chemical and Crystallographic Study on the Oxidized Binuclear Center of Cytochrome C Oxidase.
Biochim.Biophys.Acta, 1807, 2011
|
|
2YOG
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- alpha-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxol an-2-yl]methyl]thiourea, THYMIDYLATE KINASE | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
2Y80
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3UH0
 
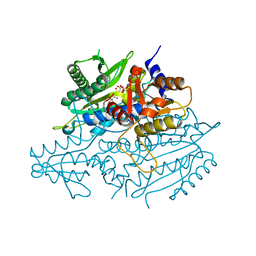 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with threonyl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UIB
 
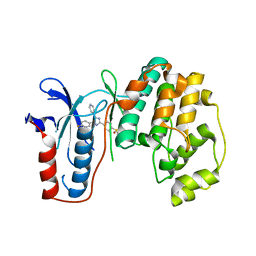 | | Map kinase LMAMPK10 from leishmania major in complex with SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, mitogen-activated protein kinase | | Authors: | Horjales, S, Schmidt-Arras, D, Leclercq, O, Spath, G, Buschiazzo, A. | | Deposit date: | 2011-11-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of the MAP Kinase LmaMPK10 from Leishmania Major Reveals Parasite-Specific Features and Regulatory Mechanisms.
Structure, 20, 2012
|
|
4NW7
 
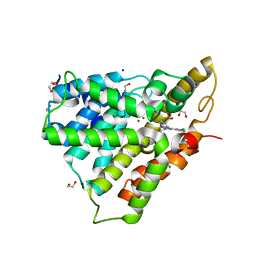 | | PDE4 catalytic domain | | Descriptor: | (4-{[4-(3-chlorophenyl)-6-cyclopropyl-1,3,5-triazin-2-yl]amino}phenyl)acetic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fox III, D, Edwards, T.E. | | Deposit date: | 2013-12-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of triazines as selective PDE4B versus PDE4D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O1J
 
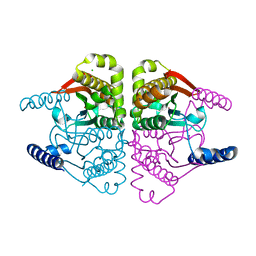 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
4O34
 
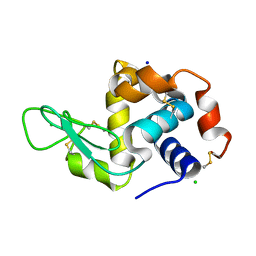 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
3UNL
 
 | | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2011-11-15 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni
To be Published
|
|
6QGA
 
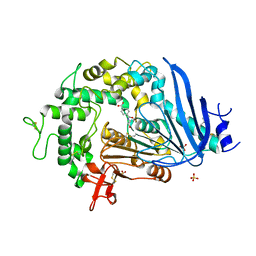 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
4O6M
 
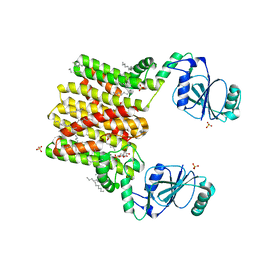 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
3UGQ
 
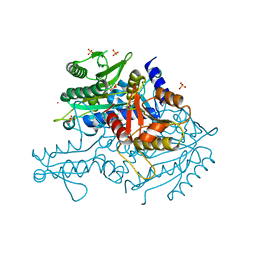 | | Crystal structure of the apo form of the yeast mitochondrial threonyl-tRNA synthetase determined at 2.1 Angstrom resolution | | Descriptor: | POTASSIUM ION, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6QGC
 
 | | PETase from Ideonella sakaiensis without ligand | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
8DXT
 
 | | Fab arm of antibody GAR12 bound to the receptor binding domain of SARS-CoV-2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab arm of antibody GAR12, Light chain of Fab arm of antibody GAR12, ... | | Authors: | Langley, D.B, Christ, D, Henry, J.Y. | | Deposit date: | 2022-08-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
3ULF
 
 | |
