8JOS
 
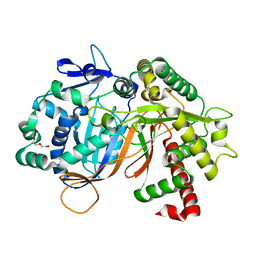 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JQ4
 
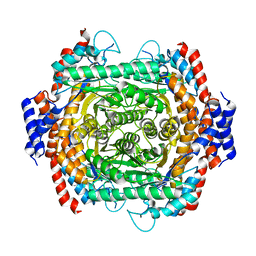 | |
8JQ3
 
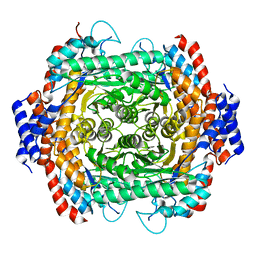 | |
8JLF
 
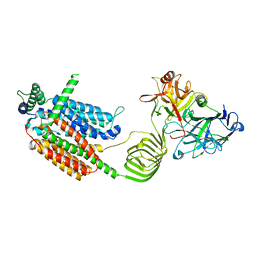 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JH8
 
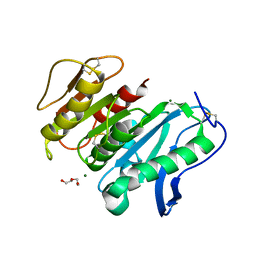 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Feruloyl esterase, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
1Y92
 
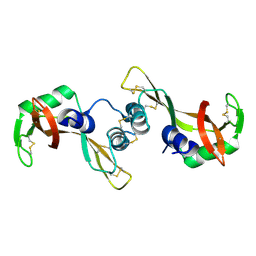 | | Crystal structure of the P19A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
1LHT
 
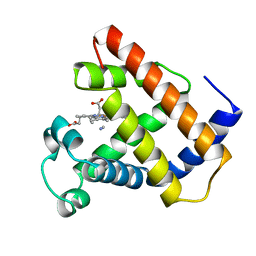 | | LOGGERHEAD SEA TURTLE MYOGLOBIN (CYANO-MET) | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Tarricone, C, Lania, A, Desideri, A, De Sanctis, G, Coletta, M, Petruzzelli, R, Ascenzi, P, Coda, A, Bolognesi, M. | | Deposit date: | 1995-02-01 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reptile heme protein structure: X-ray crystallographic study of the aquo-met and cyano-met derivatives of the loggerhead sea turtle (Caretta caretta) myoglobin at 2.0 A resolution.
J.Mol.Biol., 247, 1995
|
|
6CN0
 
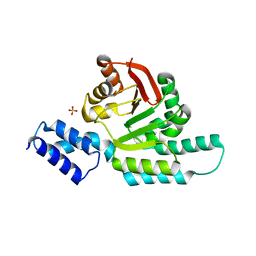 | | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Evdokimova, E, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis.
To Be Published
|
|
8JH9
 
 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Feruloyl esterase with ferulic acid, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
8AI6
 
 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI4
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
2WWE
 
 | | Crystal structure of the phox homology domain of human phosphoinositide-3-kinase-C2-gamma | | Descriptor: | PHOSPHOINOSITIDE-3-KINASE, CLASS 2, GAMMA POLYPEPTIDE | | Authors: | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the Phox Homology Domain of Human Phosphoinositide-3-Kinase-C2-Gamma
To be Published
|
|
8JLE
 
 | |
8JLG
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JS8
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8AI3
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8J9C
 
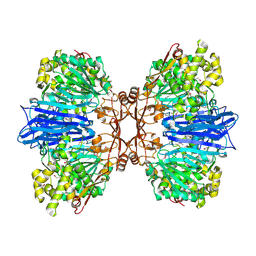 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 2024
|
|
8AI2
 
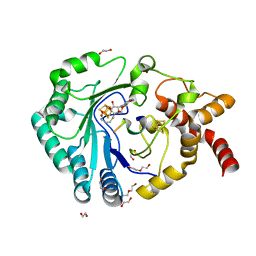 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
5K1V
 
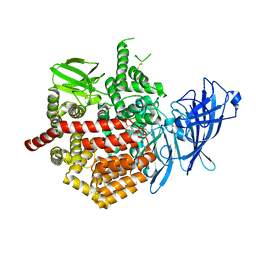 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
8J9D
 
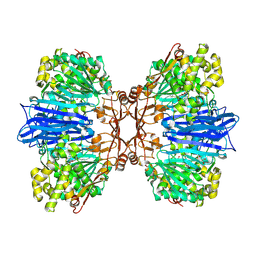 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 2024
|
|
8AI5
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8JLC
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLH
 
 | | Cryo-EM structure of SV2A dimer in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLI
 
 | | Cryo-EM structure of SV2A dimer in complex levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, Synaptic vesicle glycoprotein 2A | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
