6ALQ
 
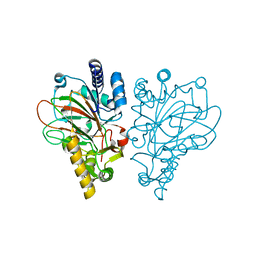 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6AMN
 
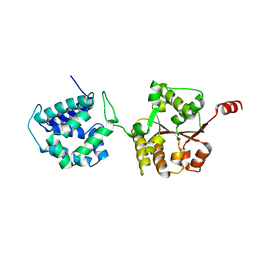 | | Crystal Structure of Hsp104 N Domain | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation.
Sci Rep, 7, 2017
|
|
6ALW
 
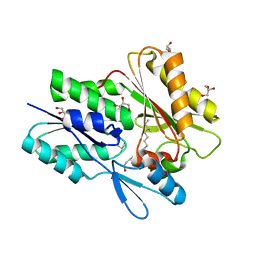 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 12-Methyl Myristic Acid (C15:0) to 1.63 Angstrom resolution | | Descriptor: | (12R)-12-methyltetradecanoic acid, (12S)-12-methyltetradecanoic acid, EDD domain protein, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6ALR
 
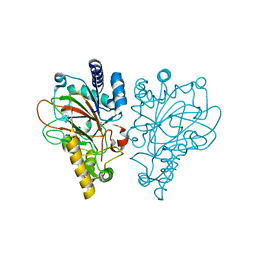 | | VioC L-arginine hydroxylase bound to the vanadyl ion, L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, SUCCINIC ACID, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Bergman, J.A, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALP
 
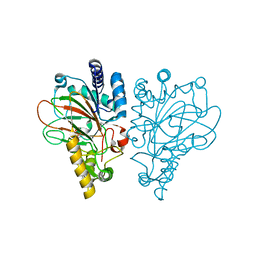 | | VioC L-arginine hydroxylase bound to Fe(II), 3S-hydroxy-L-arginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6AHZ
 
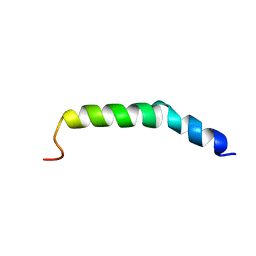 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
6B23
 
 | |
6B5X
 
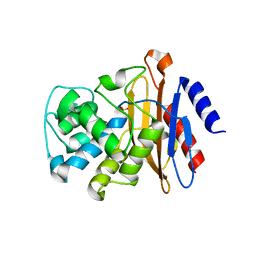 | | Beta-Lactamase, unmixed shards crystal form | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6F
 
 | |
6ANR
 
 | |
6AVD
 
 | |
6AT9
 
 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-A*0101 | | Descriptor: | ALK, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-08-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9503 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
6BFI
 
 | |
6AQ1
 
 | | The crystal structure of human FABP3 | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.40000093 Å) | | Cite: | SAR studies on truxillic acid mono esters as a new class of antinociceptive agents targeting fatty acid binding proteins.
Eur J Med Chem, 154, 2018
|
|
2PMQ
 
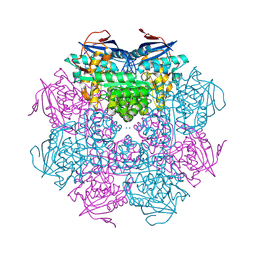 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
5J7T
 
 | |
3ZX9
 
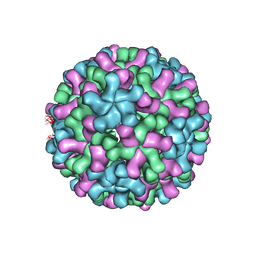 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Pearson, A.R, Stockley, P.G, Ranson, N.A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
2PZF
 
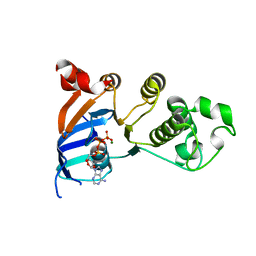 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
3ZDU
 
 | | Crystal structure of the human CDKL3 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-DEPENDENT KINASE-LIKE 3, SODIUM ION, ... | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Pike, A.C.W, Quigley, A, MacKenzie, A, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
2PYB
 
 | |
5J1B
 
 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - WT. | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Maveyraud, L, Batista, M, Martin-yken, H, Francois, J.M, Zerbib, D, Mourey, L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biophysical characterization of Knr4, an intrinsically disordered hub protein form Saccharomyces cerevisiae
To Be Published
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q7C
 
 | | Crystal structure of IQN17 | | Descriptor: | CHLORIDE ION, fusion protein between yeast variant GCN4 and HIVgp41 | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Kim, P.S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
2Q3I
 
 | | Crystal structure of the D10-P3/IQN17 complex: a D-peptide inhibitor of HIV-1 entry bound to the GP41 coiled-coil pocket | | Descriptor: | CHLORIDE ION, D-peptide, Fusion protein between the Coiled-Coil pocket of HIV GP41 and gcn4-PIQI | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
5HI5
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (4S,20R)-7-chloro-N-methyl-4-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-3,18-dioxo-2,19-diazatetracyclo[20.2.2.1~6,10~.1~11,15~]octacosa-1(24),6(28),7,9,11(27),12,14,22,25-nonaene-20-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
