8HM1
 
 | |
8HM3
 
 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM2
 
 | |
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8J37
 
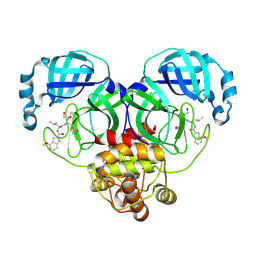 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8HIW
 
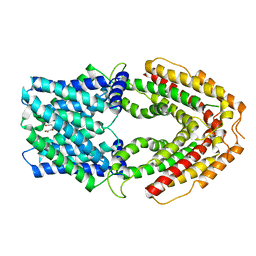 | | AtALMT9 in the apo state | | Descriptor: | Aluminum-activated malate transporter 9, CITRIC ACID | | Authors: | Gong, D.S. | | Deposit date: | 2022-11-22 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insight into the Arabidopsis vacuolar anion channel ALMT9 shows clade specificity.
Cell Rep, 43, 2024
|
|
8J3A
 
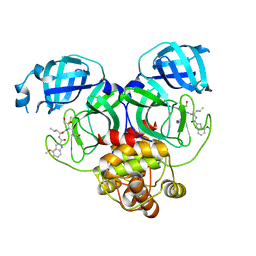 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8HIY
 
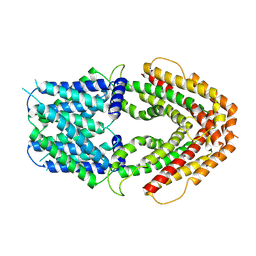 | | AtALMT9 plus malate | | Descriptor: | Aluminum-activated malate transporter 9 | | Authors: | Gong, D.S. | | Deposit date: | 2022-11-22 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into the Arabidopsis vacuolar anion channel ALMT9 shows clade specificity.
Cell Rep, 43, 2024
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | Descriptor: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-05-01 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8XTC
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, GLYCEROL, umgsp2-mut | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
8JYA
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYF
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with DMAPP | | Descriptor: | Butyrophylin 3, DIMETHYLALLYL DIPHOSPHATE, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JY9
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYB
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 | | Descriptor: | Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
4N0S
 
 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
6IVY
 
 | | Crystal structure of iron-bound HitA from Pseudomonas aeruginosa | | Descriptor: | FE (III) ION, PHOSPHATE ION, Periplasmic Ferric iron-binding Protein HitA | | Authors: | Zhang, Z.R, Li, H.Y. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9999994 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
4RAX
 
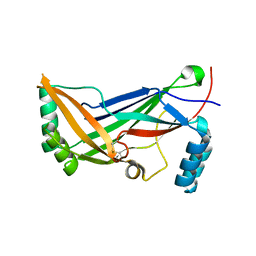 | | A regulatory domain of an ion channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Yang, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
9J3M
 
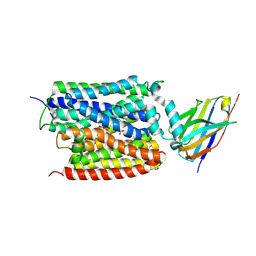 | | ADP/Pi bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3L
 
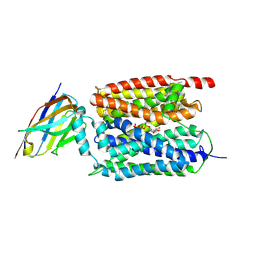 | | ATP bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
