6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
5WDF
 
 | |
5TE4
 
 | | Crystal Structure of Broadly Neutralizing VRC01-class Antibody N6 in Complex with HIV-1 Clade G Strain X2088 gp120 Core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth.
Immunity, 45, 2016
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TO7
 
 | | Cryo-EM structure of HERH-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HERH-b*01 heavy chain, ... | | Authors: | Roark, R.S, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
5TE6
 
 | |
5TE7
 
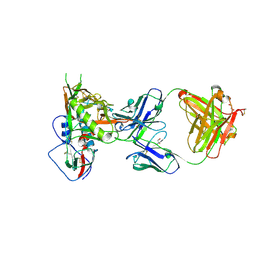 | |
8ZF4
 
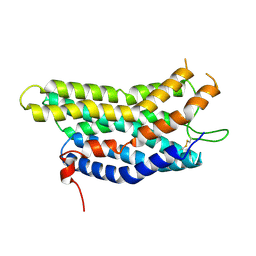 | |
8ZFB
 
 | |
8ZF7
 
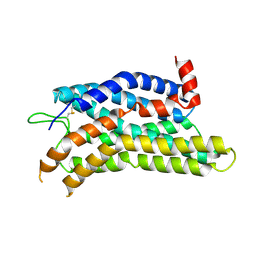 | |
8ZFC
 
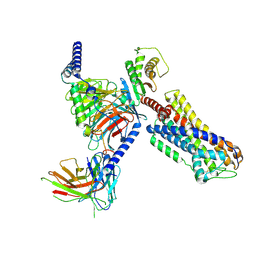 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.6 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFE
 
 | |
8ZFA
 
 | | Cryo-EM structure of the xtGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF9
 
 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZD1
 
 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-04-30 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF6
 
 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.7 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFD
 
 | |
6KVM
 
 | | Crystal structure of Chicken MHC Class II for 1.9 angstrom | | Descriptor: | GLYCEROL, MHC class II alpha chain, MHC class II beta chain 2, ... | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Newly Recognized Pairing Mechanism of the alpha- and beta-Chains of the Chicken Peptide-MHC Class II Complex.
J Immunol., 204, 2020
|
|
7RI2
 
 | |
7RI1
 
 | |
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7Y89
 
 | | Structure of the GPR17-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Ye, F, Chen, G. | | Deposit date: | 2022-06-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of G-protein-coupled receptor GPR17 in complex with inhibitory G protein.
MedComm (2020), 3, 2022
|
|
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
