3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
7CYN
 
 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4GEN
 
 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEM
 
 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
1UDM
 
 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
6LN5
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN9
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class2) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN7
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN8
 
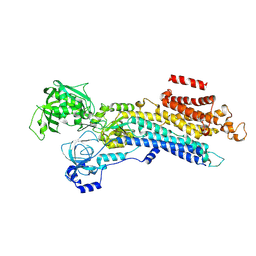 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class1) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LLE
 
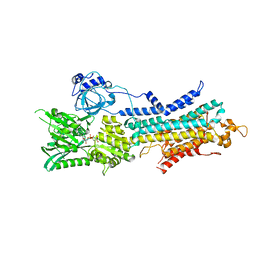 | | CryoEM structure of SERCA2b WT in E1-2Ca2+-AMPPCP state. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LLY
 
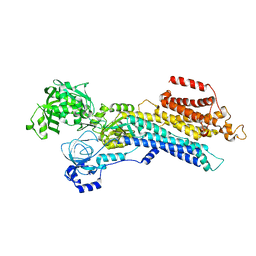 | | CryoEM structure of SERCA2b WT in E2-BeF3- state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
7E7Z
 
 | |
7E83
 
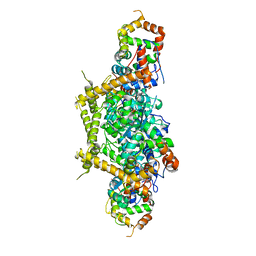 | |
7E87
 
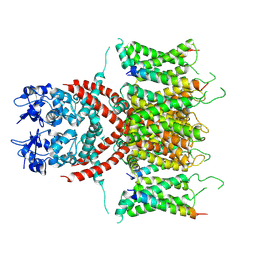 | |
7E84
 
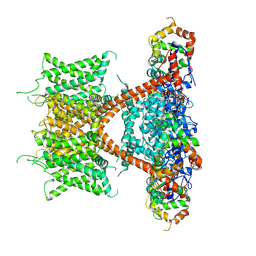 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-02-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8H
 
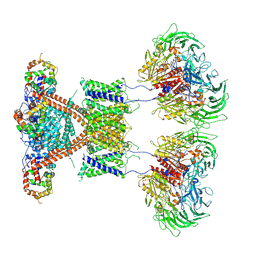 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8E
 
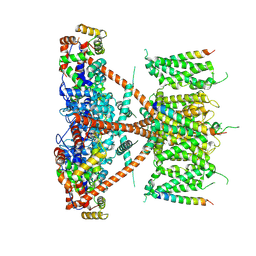 | |
7E8G
 
 | |
7E89
 
 | |
7E8B
 
 | | CryoEM structure of human Kv4.2-DPP6S complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7VSG
 
 | |
7VSH
 
 | |
6LN6
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class2) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
7F3F
 
 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F0J
 
 | | CryoEM structure of human Kv4.2 | | Descriptor: | Potassium voltage-gated channel subfamily D member 2, ZINC ION | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
