9F9K
 
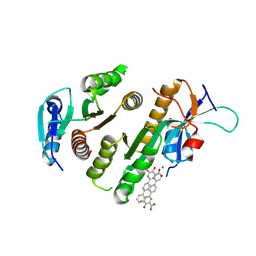 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F99
 
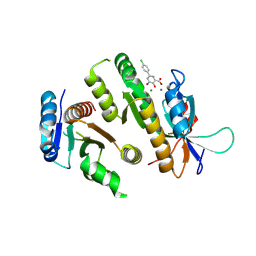 | | Crystal structure of MUS81-EME1 bound by compound 10. | | Descriptor: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
2DUF
 
 | |
2DUI
 
 | |
2DUE
 
 | |
2DUH
 
 | |
2DUG
 
 | |
7DRV
 
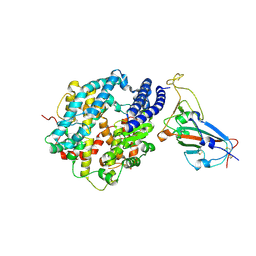 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
5APJ
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5APK
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR ROR-GAMMA | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5APH
 
 | | Ligand complex of RORg LBD | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-FLUOROPHENYL)-4-[(4-FLUOROPHENYL)SULFONYL]-2,3,4,5-TETRAHYDRO-1,4-BENZOXAZEPIN-6-AMINE, NUCLEAR RECEPTOR COACTIVATOR 2, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
7C8D
 
 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
7DDP
 
 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8XOM
 
 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOL
 
 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJS
 
 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
4DDS
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 11 | | Descriptor: | 3-(pyrimidin-2-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4DE0
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 16 | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, N-[3-(1H-tetrazol-5-yl)phenyl]-1H-benzimidazole-7-carboxamide | | Authors: | Nichols, D.N, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
