7FBI
 
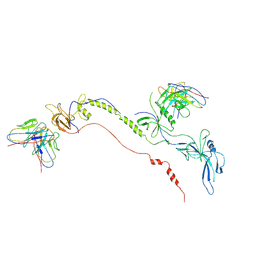 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
1EV7
 
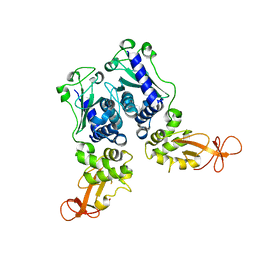 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
4IWM
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in P21 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
8EHK
 
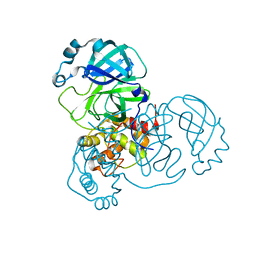 | |
8EHL
 
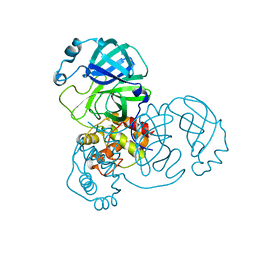 | |
8EHM
 
 | |
8EHJ
 
 | |
6BGJ
 
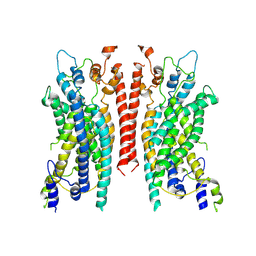 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
1FGL
 
 | | Cyclophilin A complexed with a fragment of HIV-1 GAG protein | | Descriptor: | CYCLOPHILIN A, HIV-1 GAG PROTEIN | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1996-11-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity.
Structure, 5, 1997
|
|
4IWG
 
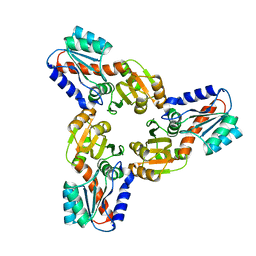 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
1MN9
 
 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
6XD7
 
 | | KPC-2 N170A mutant bound to hydrolyzed ampicillin at 1.65 A | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
6Y8J
 
 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
1M6X
 
 | | Flpe-Holliday Junction Complex | | Descriptor: | Flp recombinase, Symmetrized FRT site | | Authors: | Conway, A.B, Chen, Y, Rice, P.A. | | Deposit date: | 2002-07-17 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Plasticity of the Flp-Holliday Junction Complex
J.Mol.Biol., 326, 2003
|
|
6XD5
 
 | | Apo KPC-2 N170A mutant at 1.20 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
6Y9D
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y8S
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6XJ8
 
 | | KPC-2 N170A mutant bound to hydrolyzed imipenem at 2.05 A | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
2IE4
 
 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | Descriptor: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
1NYK
 
 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
1MN7
 
 | | NDP kinase mutant (H122G;N119S;F64W) in complex with aBAZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXY-THYMIDINE-5'-ALPHA BORANO TRIPHOSPHATE, MAGNESIUM ION, NDP kinase | | Authors: | gallois-montbrun, s, schneider, b, chen, y, giacomoni-fernandes, v, mulard, l, morera, s, janin, j, deville-bonne, d, veron, m. | | Deposit date: | 2002-09-05 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation
J.BIOL.CHEM., 277, 2002
|
|
1TM0
 
 | | Crystal Structure of the putative proline racemase from Brucella melitensis, Northeast Structural Genomics Target LR31 | | Descriptor: | PROLINE RACEMASE | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Ho, C.K, Ma, L.-C, Cooper, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2NPP
 
 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
1P4E
 
 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
