6A4X
 
 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A5G
 
 | |
6A5H
 
 | |
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
4I3R
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HDS
 
 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
 | |
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4JAN
 
 | | crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6JPM
 
 | |
4JAM
 
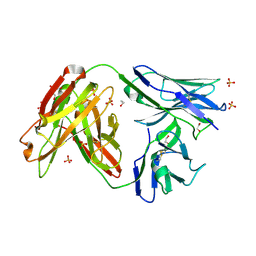 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | Descriptor: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
4I3S
 
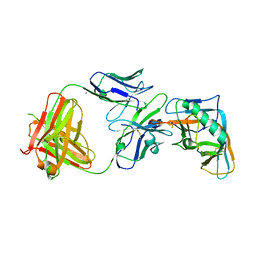 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 | | Descriptor: | CALCIUM ION, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
7F02
 
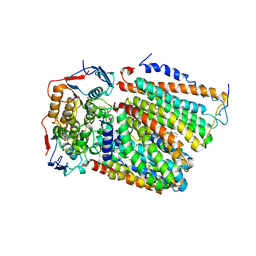 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
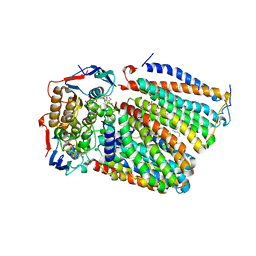 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6LY4
 
 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8TS6
 
 | | Cyanophage A-1(L) portal | | Descriptor: | Portal protein | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
7FE6
 
 | |
