8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKI
 
 | | Cryo-EM structure of the distal rod-hook within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-27 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP1
 
 | | Cryo-EM structure of the protomers of the C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP0
 
 | | Cryo-EM structure of the protomers of the C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
6N80
 
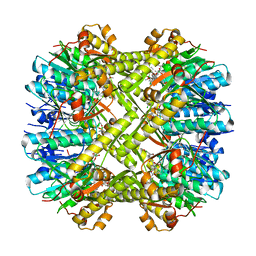 | | S. aureus ClpP bound to anti-4a | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(1R)-1-borono-3-methylbutyl]-N~2~-(2-chloro-4-methoxybenzene-1-carbonyl)-L-leucinamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-11-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | De Novo Design of Boron-Based Peptidomimetics as Potent Inhibitors of Human ClpP in the Presence of Human ClpX.
J.Med.Chem., 62, 2019
|
|
7CG7
 
 | | Cryo-EM structure of the flagellar MS ring with C34 symmetry from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBL
 
 | | Cryo-EM structure of the flagellar LP ring from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGO
 
 | | Cryo-EM structure of the flagellar motor-hook complex from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar MS ring L1, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGB
 
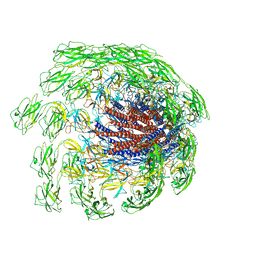 | | Cryo-EM structure of the flagellar hook from Salmonella | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
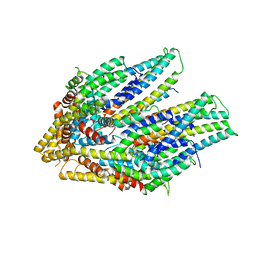 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBM
 
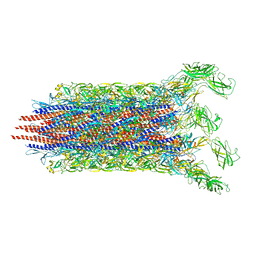 | | Cryo-EM structure of the flagellar distal rod with partial hook from Salmonella | | Descriptor: | Flagellar basal-body rod protein FlgF, Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
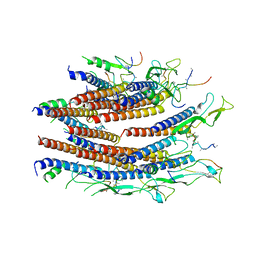 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E80
 
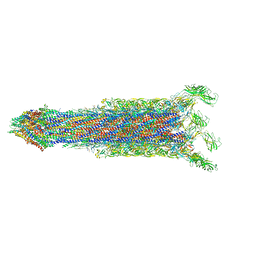 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E81
 
 | | Cryo-EM structure of the flagellar MS ring with FlgB-Dc loop and FliE-helix 1 from Salmonella | | Descriptor: | Flagellar M-ring protein, FlgB-Dc loop, FliE helix 1 | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
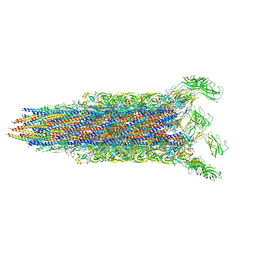 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
8CC4
 
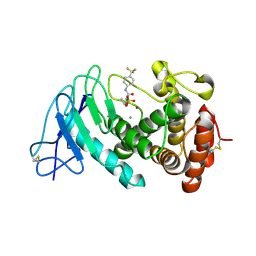 | | LasB bound to phosphonic acid based inhibitor | | Descriptor: | CALCIUM ION, Elastase, ZINC ION, ... | | Authors: | Mueller, R, Sikandar, A. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections.
Acs Cent.Sci., 9, 2023
|
|
7XMH
 
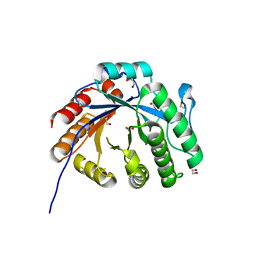 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
6WRP
 
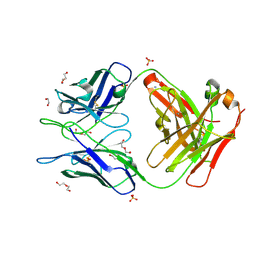 | | Crystal Structure of PI3-E12 Fab, An Antibody Against Human Parainfluenza Virus Type III | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Weidle, C, Pancera, M. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Protective antibodies against human parainfluenza virus type 3 infection.
Mabs, 13, 2021
|
|
7Y86
 
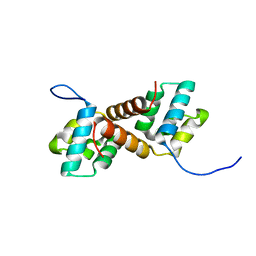 | | CcpS mutant | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
7Y8Z
 
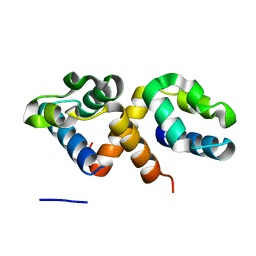 | | CcpS | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
5J78
 
 | | Crystal structure of an Acetylating Aldehyde Dehydrogenase from Geobacillus thermoglucosidasius | | Descriptor: | ACETATE ION, Acetaldehyde dehydrogenase (Acetylating), GLYCEROL, ... | | Authors: | Crennell, S.J, Extance, J.P, Danson, M.J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius.
Protein Sci., 25, 2016
|
|
5J7I
 
 | |
5TUQ
 
 | | Crystal Structure of a 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[(benzyloxy)methyl]-6-(cyclohexylmethyl)-3-hydroxy-5-(propan-2-yl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverase transcriptase: Impacts of the 3-OH on inhibiting RNase H and polymerase.
Eur J Med Chem, 128, 2017
|
|
2LVM
 
 | |
