7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
5YWO
 
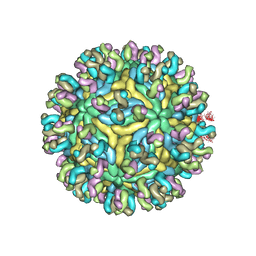 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-09-12 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWF
 
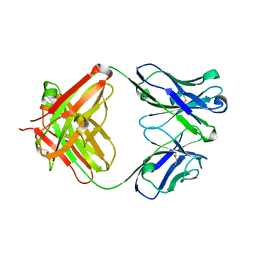 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
8EWW
 
 | |
8EY1
 
 | |
8EY9
 
 | | Structure of Arabidopsis fatty acid amide hydrolase mutant S305A in complex with 9-hydroxy-10,12-octadecadienoyl-ethanolamide | | Descriptor: | (9R,10E,12Z)-9-hydroxy-N-(2-hydroxyethyl)octadeca-10,12-dienamide, Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Gaguancela, O.A, Chapman, K.D. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural interactions explain the versatility of FAAH in the hydrolysis of plant and microbial acyl amide signals
To be published
|
|
4ZP0
 
 | | Crystal structure of E. coli multidrug transporter MdfA in complex with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
4ZOW
 
 | | Crystal structure of E. coli multidrug transporter MdfA in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
4ZP2
 
 | | Crystal structure of E. coli multidrug transporter MdfA in complex with n-dodecyl-N,N-dimethylamine-N-oxide | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
6J0B
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath-tube complex in extended state | | Descriptor: | Pvc1, Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0C
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath complex in contracted state | | Descriptor: | Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0F
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath/tube terminator in extended state | | Descriptor: | Pvc1, Pvc16 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
3DAN
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3DBM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | (9E,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-06-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6J0M
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC baseplate in extended state (reconstructed with C3 symmetry) | | Descriptor: | Pvc8 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0N
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, baseplate in extended state, refined in C6 symmetry | | Descriptor: | Pvc1, Pvc11, Pvc12, ... | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
5BYY
 
 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ABJ
 
 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
3DAM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5BYZ
 
 | | ERK5 in complex with small molecule | | Descriptor: | 4-({5-fluoro-4-[2-methyl-1-(propan-2-yl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)-N-[2-(piperidin-1-yl)ethyl]benzamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4QDJ
 
 | |
4QDK
 
 | |
