7VQC
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with pyrophosphate | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, PYROPHOSPHATE 2-, SULFATE ION | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQD
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and geranylgeranyl pyrophosphate. | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, NerylNeryl pyrophosphate, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQA
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQ9
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl thiopyrophosphate and isopentyl S-thiolodiphosphate | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7F3P
 
 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
5E6V
 
 | |
6M6O
 
 | | NMR SOLUTION STRUCTURE OF A C-FLIPs | | Descriptor: | CASP8 and FADD-like apoptosis regulator | | Authors: | Bai, Z.Q, Hu, K.F. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of c-FLIP death effector domains.
Biochem.Biophys.Res.Commun., 617, 2022
|
|
8JTR
 
 | | Cryo-EM structure of GeoCas9-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA (139-bp) | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-22 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of Geobacillus stearothermophilus Cas9: Insights into the Catalytic Process and Thermostability of CRISPR-Cas9
Acs Catalysis, 14, 2024
|
|
8E3D
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#11) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*AP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*AP*T)-3'), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
4Z7P
 
 | | X-ray structure of racemic ShK Q16K toxin | | Descriptor: | Potassium channel toxin kappa-stichotoxin-She1a, SULFATE ION | | Authors: | Sickmier, E.A. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Pharmaceutical Optimization of Peptide Toxins for Ion Channel Targets: Potent, Selective, and Long-Lived Antagonists of Kv1.3.
J.Med.Chem., 58, 2015
|
|
8JTJ
 
 | | Cryo-EM structure of GeoCas9-sgRNA-dsDNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (29-MER), DNA (5'-D(P*GP*GP*GP*CP*GP*CP*GP*AP*A)-3'), ... | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of Geobacillus stearothermophilus Cas9: Insights into the Catalytic Process and Thermostability of CRISPR-Cas9
Acs Catalysis, 14, 2024
|
|
8E3E
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#10) | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*A)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8ZY2
 
 | | Sarbecovirus BANAL-20-52 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY9
 
 | | Ra9479 Bat ACE2 Dimer in Complex with Two BtKY72 Sarbecovirus Spike RBDs. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY7
 
 | | Sarbecovirus HeB2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY4
 
 | | Sarbecovirus YN2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY6
 
 | | Sarbecovirus GX2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY3
 
 | | Sarbecovirus BANAL-20-236 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY1
 
 | | Sarbecovirus BM48-31 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZYA
 
 | | Ra9479 Bat ACE2 Bound to BtkY72 Sarbecovirus Spike RBD (Focused Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY5
 
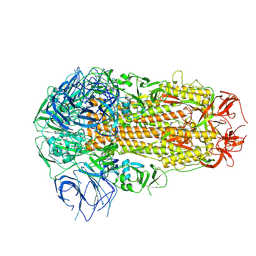 | | Sarbecovirus RmYN02 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY0
 
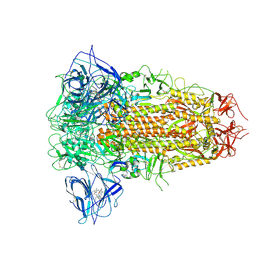 | | Sarbecovirus BtKY72 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8SSU
 
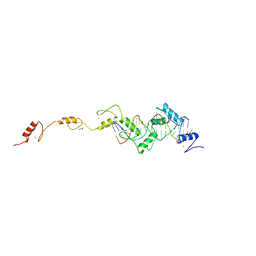 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
