3FSH
 
 | |
5U5T
 
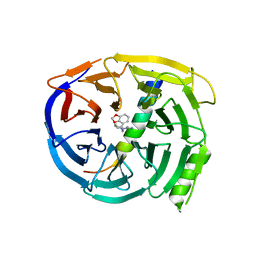 | | Crystal structure of EED in complex with H3K27Me3 peptide and 3-(benzo[d][1,3]dioxol-4-ylmethyl)piperidine-1-carboximidamide | | Descriptor: | (3R)-3-[(2H-1,3-benzodioxol-4-yl)methyl]piperidine-1-carboximidamide, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5UEX
 
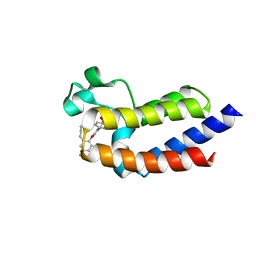 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5U5K
 
 | |
5UEU
 
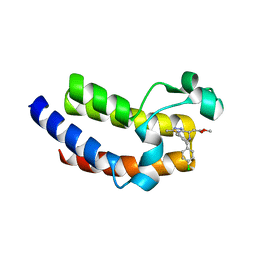 | | BRD4_BD2_A-1107604 | | Descriptor: | Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEZ
 
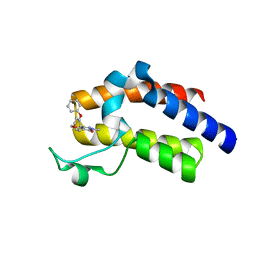 | | BRD4_BD2_A-1342843 | | Descriptor: | 5-methoxy-2-methyl-6-(2-phenoxyphenyl)pyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UF0
 
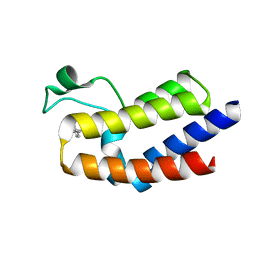 | | BRD4_BD2-A-35165 | | Descriptor: | 2-methyl-5-(methylamino)-6-phenylpyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VLR
 
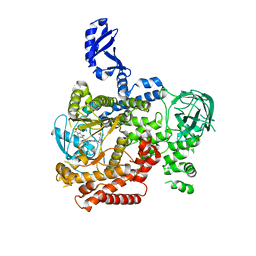 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A TRIFLUORO-ETHYL-PYRAZOL-PYROLOTRIAZINE INHIBITOR | | Descriptor: | 4-acetyl-1-(3-{4-amino-5-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)-3,3-dimethylpiperazin-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
J. Med. Chem., 60, 2017
|
|
5U5H
 
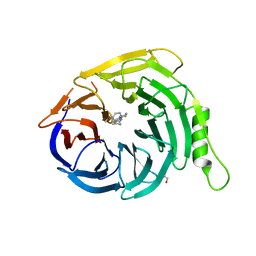 | | Crystal structure of EED in complex with 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2-fluoro-5-methoxyphenyl)methyl]-1-(propan-2-yl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Polycomb protein EED | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5U62
 
 | | Crystal structure of EED in complex with H3K27Me3 peptide and 6-(benzo[d][1,3]dioxol-4-ylmethyl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2H-1,3-benzodioxol-4-yl)methyl]-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
7VFA
 
 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFB
 
 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7FCP
 
 | |
7FCQ
 
 | |
5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEY
 
 | | BRD4_BD2_A-1412838 | | Descriptor: | 5-[2-(2,4-difluorophenoxy)-5-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}phenyl]-4-ethoxy-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
1AWY
 
 | | NMR STRUCTURE OF CALCIUM BOUND CONFORMER OF CONANTOKIN G, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CONANTOXIN G | | Authors: | Rigby, A.C, Baleja, J.D, Leping, L, Pedersen, L.G, Furie, B.C, Furie, B. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus.
Biochemistry, 36, 1997
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
8HLO
 
 | | Crystal structure of ASAP1-SH3 and MICAL1-PRM complex | | Descriptor: | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, Proline rich motif from MICAL1, ... | | Authors: | Niu, F, Wei, Z, Yu, C. | | Deposit date: | 2022-11-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Int J Mol Sci, 24, 2023
|
|
2R7I
 
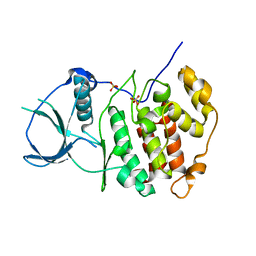 | | Crystal structure of catalytic subunit of protein kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
2R6M
 
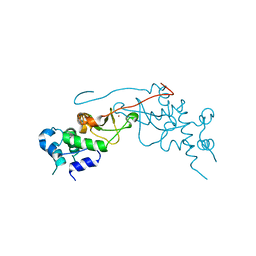 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
3SV5
 
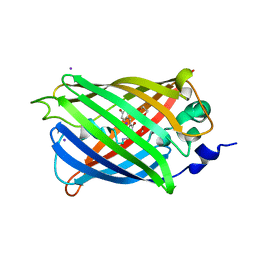 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
6DEH
 
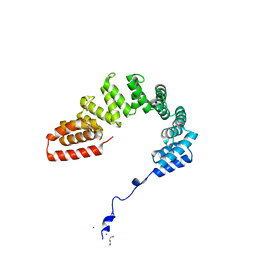 | | Structure of LpnE Effector Protein from Legionella pneumophila (sp. Philadelphia) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Voth, K, Chung, I.Y.W, van Straaten, K.E, Cygler, M. | | Deposit date: | 2018-05-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein.
FEBS J., 286, 2019
|
|
7KBB
 
 | |
7KBA
 
 | |
