6J8F
 
 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J7N
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7S
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) wild type protein from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J8N
 
 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J9H
 
 | |
6J7P
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.629 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
8H7B
 
 | | The crystal structure of human mcl1 kinase domain in complex with MCL1-M-EBA | | Descriptor: | 7-[3-(isoquinolin-7-yloxymethyl)-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46408451 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
8H7F
 
 | | The crystal structure of human abl1 kinase domain in complex with abl1-B-EBA | | Descriptor: | 1-[6-(6-methoxyisoquinolin-7-yl)-1,3-benzothiazol-2-yl]-3-(2-oxidanylideneethyl)urea, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45013428 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
8H7H
 
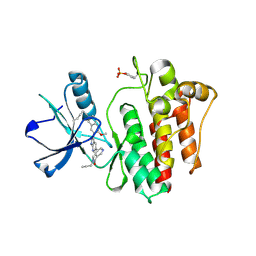 | | The crystal structure of human abl1 kinase domain in complex with abl1-A-EBA | | Descriptor: | 5-[3-(6-methoxyisoquinolin-7-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-methyl-N-prop-2-ynyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27789712 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
3T7K
 
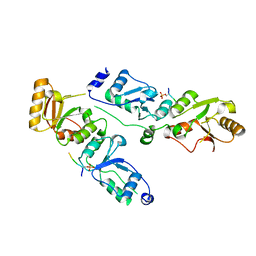 | | Complex structure of Rtt107p and phosphorylated histone H2A | | Descriptor: | Histone H2A.1, Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
8XM1
 
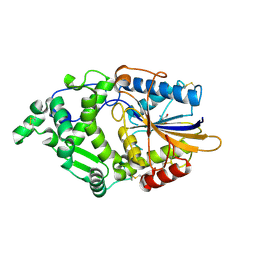 | | Phytase mutant APPAmut4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phytase | | Authors: | Tu, T, Wang, Q. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Achieving thermostability of a phytase with resistance up to 100 °C.
J.Biol.Chem., 300, 2024
|
|
8XSF
 
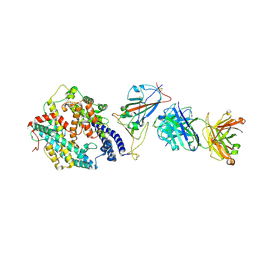 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSI
 
 | | SARS-CoV-2 RBD + IMCAS-364 (Local Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-364 H chain, IMCAS-364 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8Y0Y
 
 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | Descriptor: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSJ
 
 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSL
 
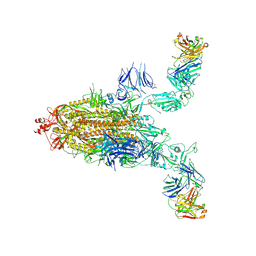 | | SARS-CoV-2 spike + IMCAS-123 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 heavy chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XM2
 
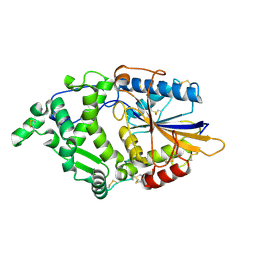 | |
3T7I
 
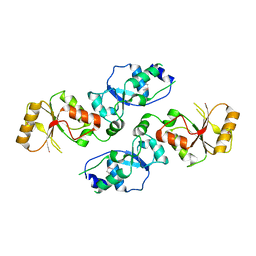 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6J7Q
 
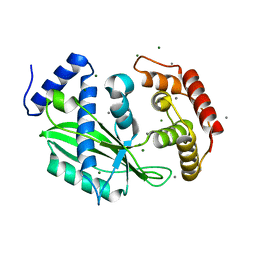 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A from Mycobacterium tuberculosis | | Descriptor: | CALCIUM ION, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J91
 
 | | Structure of a hypothetical protease | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
3T7J
 
 | | Crystal structure of Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
7WH8
 
 | |
7WHB
 
 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
