5YC7
 
 | |
6KY5
 
 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
9JE9
 
 | | Crystal structure of a amidase that can hydrolase PU plastic | | Descriptor: | Amidase | | Authors: | Li, Z.S, Fang, S.T, Zheng, Z.R, Xia, W, Zhou, H.Y, Li, W.S, You, S, Han, X, Liu, W.D. | | Deposit date: | 2024-09-02 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Engineering of a Urethanase for Enhanced Depolymerization of Polyurethane
Acs Catalysis, 15, 2025
|
|
8XTC
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, GLYCEROL, umgsp2-mut | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
8XTB
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, umgsp2 | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Wei, R, Liu, W.D, Uwe, B. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium
To Be Published
|
|
8XRZ
 
 | | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis | | Descriptor: | 4-oxidanylbutyl ~{N}-[4-[(4-aminophenyl)methyl]phenyl]carbamate, Carboxylic ester hydrolase, SULFATE ION | | Authors: | Li, Z.S, Wang, H, Zheng, Z.R, Cong, L, Chen, Y.Y, Han, X, Wei, R, Uwe, B, liu, W.D. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis
To Be Published
|
|
5M00
 
 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
8YMA
 
 | | CRYSTAL STRUCTURE OF A NOVEL PU PLASTIC DEGRADATION ENZYME FROM THERMAEROBACTER MARIANENSIS | | Descriptor: | Carboxylic ester hydrolase, SULFATE ION | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Han, X, Wei, R, Bornscheuer, U.T, Liu, W.D. | | Deposit date: | 2024-03-08 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | CRYSTAL STRUCTURE OF A NOVEL PU PLASTIC DEGRADATION ENZYME FROM THERMAEROBACTER MARIANENSIS
To Be Published
|
|
7ARM
 
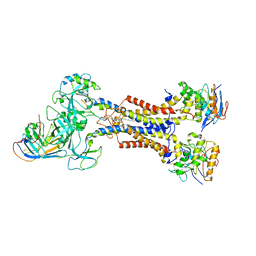 | | LolCDE in complex with lipoprotein and LolA | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
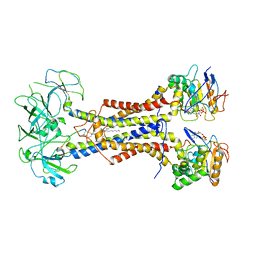 | | LolCDE in complex with lipoprotein and ADP | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARI
 
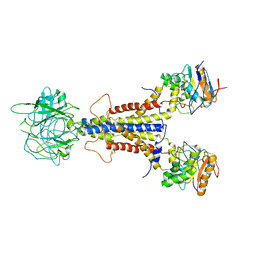 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
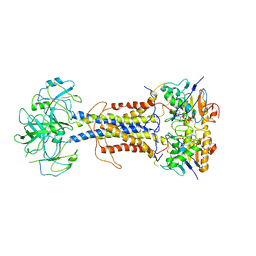 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARH
 
 | | LolCDE in complex with lipoprotein | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARJ
 
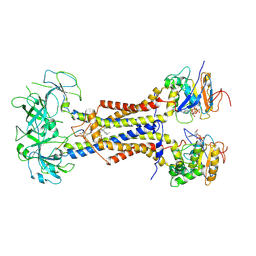 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8ZQ7
 
 | |
1NP5
 
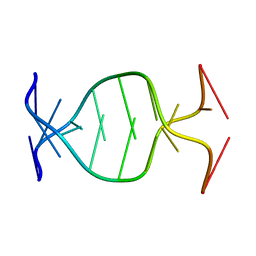 | | (GAC)3 parallel duplex | | Descriptor: | 5'-D(*GP*AP*CP*GP*AP*CP*GP*AP*C)-3' | | Authors: | Zheng, M, Han, X, Gao, X. | | Deposit date: | 2003-01-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Strand polarity of trinucleotide repeat sequences:
NMR studies of parallel/anti-pararell duplex,{d(GAC)3}2
To be Published
|
|
8IJT
 
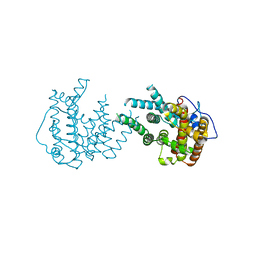 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
8II9
 
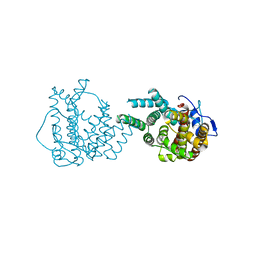 | | crystal structure of Hyp mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Liu, W.D, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Su, L.Q. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of Hyp
to be published
|
|
1CX5
 
 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
8WDW
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2, ... | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
9IK1
 
 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
