8GSY
 
 | |
8GSX
 
 | |
3CV9
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R73A/R84A mutant) in complex with 1alpha,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3CV8
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
2I57
 
 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
2HCV
 
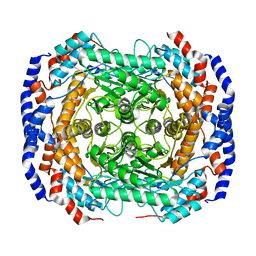 | | Crystal structure of L-rhamnose isomerase from Pseudomonas stutzeri with metal ion | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-06-19 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
2I56
 
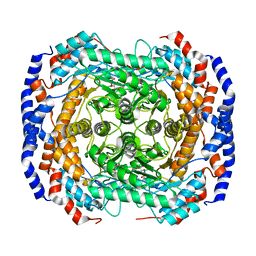 | | Crystal structure of L-Rhamnose Isomerase from Pseudomonas stutzeri with L-Rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
3IL8
 
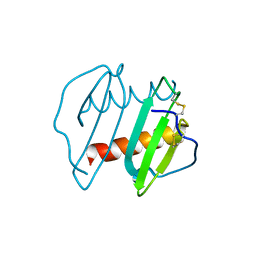 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2OHD
 
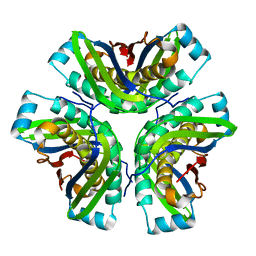 | | Crystal structure of hypothetical molybdenum cofactor biosynthesis protein C from Sulfolobus tokodaii | | Descriptor: | Probable molybdenum cofactor biosynthesis protein C | | Authors: | Yoshida, H, Yamada, M, Kuramitsu, S, Kamitori, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative molybdenum-cofactor biosynthesis protein C (MoaC) from Sulfolobus tokodaii (ST0472)
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2OU4
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii | | Descriptor: | D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-02-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUL
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii at 1.79 A resolution | | Descriptor: | D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
1WD9
 
 | | Calcium bound form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type IV, SULFATE ION | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD8
 
 | | Calcium free form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | Protein-arginine deiminase type IV | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
7WMC
 
 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7WMT
 
 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
1ZZE
 
 | | X-ray Structure of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor | | Descriptor: | Aldehyde reductase II, SULFATE ION | | Authors: | Kamitori, S, Iguchi, A, Ohtaki, A, Yamada, M, Kita, K. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS5
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2DEW
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal tail including Arg8 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEX
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal peptide including Arg17 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
