7VSJ
 
 | |
5YUH
 
 | |
5YUG
 
 | | AtVAL1 PHD-Like domain in the P31 space group | | Descriptor: | B3 domain-containing transcription repressor VAL1, GLYCEROL, ZINC ION | | Authors: | Zhang, M.M, Wu, B.X. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | AtVAL1 PHD-Like domain in the P31 space group
To Be Published
|
|
5WWG
 
 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWE
 
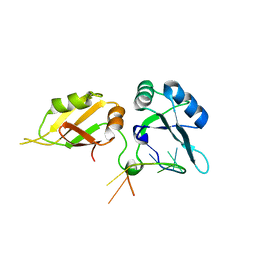 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*GP*AP*CP*UP*AP*GP*A)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWF
 
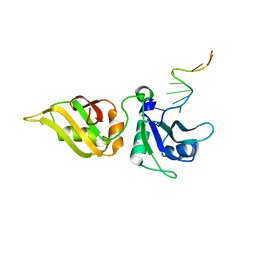 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
7VKI
 
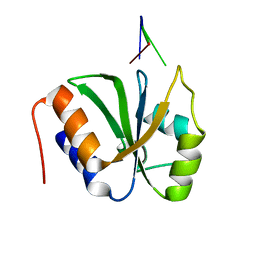 | | ESRP1 qRRM2 in complex with 12mer-RNA | | Descriptor: | Epithelial splicing regulatory protein 1, RNA (12-mer) | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Nucleic Acids Res., 2023
|
|
6J23
 
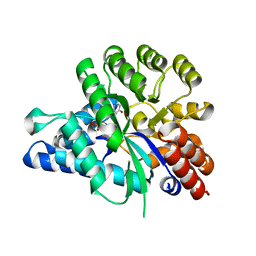 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
8KA4
 
 | |
8KA5
 
 | |
8KA3
 
 | |
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
5WSP
 
 | | Crystal structure of DNA3 duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), STRONTIUM ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSR
 
 | | Crystal structure of T-Hg-T pair containing DNA duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSQ
 
 | | Crystal structure of C-Hg-T pair containing DNA duplex | | Descriptor: | 1,3-PROPANDIOL, DNA (5'-D(*GP*CP*CP*CP*GP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSS
 
 | |
8Z2O
 
 | | Crystal structure of 5-N-alpha-glycinylthymidine (N-alpha-GlyT) FAD-dependent lyase gp47/NGTO from Pseudomonads phage PaMx11 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent lyase, ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z79
 
 | |
8Z2M
 
 | |
8Z2N
 
 | | Crystal structure of 5-phosphomethyl-2'-deoxyuridine (5-PmdU) glycinyltransferase gp46/PUGT from Pseudomonads phage PaMx11 in complex with dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*GP*TP*CP*GP*AP*CP*GP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*GP*TP*CP*GP*AP*CP*TP*A)-3'), ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
5UVG
 
 | | Crystal structure of the human neutral sphingomyelinase 2 (nSMase2) catalytic domain with insertion deleted and calcium bound | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase 3,Sphingomyelin phosphodiesterase 3 | | Authors: | Airola, M.V, Guja, K.E, Garcia-Diaz, M, Hannun, Y.A. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structure of human nSMase2 reveals an interdomain allosteric activation mechanism for ceramide generation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IS5
 
 | |
8IS4
 
 | | Structure of an Isocytosine specific deaminase Vcz in complexed with 5-FU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Guo, W.T, Li, X.J, Wu, B.X. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
