3UQN
 
 | | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, OXAMIC ACID | | Authors: | Singh, A, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution
To be Published
|
|
3TAK
 
 | | Crystal structure of the complex of DHDPS from Acinetobacter baumannii with Pyruvate at 1.4 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Singh, A, Kaushik, S, Sinha, M, Tewari, R, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-04 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of the complex of DHDPS from Acinetobacter baumannii with Pyruvate at 1.4 A resolution
To be Published
|
|
4N42
 
 | | Crystal structure of allergen protein scam1 from Scadoxus multiflorus | | Descriptor: | PHOSPHATE ION, Xylanase and alpha-amylase inhibitor protein isoform III | | Authors: | Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of allergen protein scam1 from Scadoxus multiflorus
To be published
|
|
3O9N
 
 | | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Singh, A, Kumar, S, Sinha, M, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution
To be Published
|
|
5GQO
 
 | |
1PTJ
 
 | | Crystal structure analysis of the DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
1PT9
 
 | | Crystal Structure Analysis of the DIII Component of Transhydrogenase with a Thio-Nicotinamide Nucleotide Analogue | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
5ZRG
 
 | | M. smegmatis antimutator protein MutT2 in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRK
 
 | | M. smegmatis antimutator protein MutT2 in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRP
 
 | | M. smegmatis antimutator protein MutT2 form 3 | | Descriptor: | Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRI
 
 | | M. smegmatis antimutator protein MutT2 in complex with 5m-dCMP | | Descriptor: | MAGNESIUM ION, Putative mutator protein MutT2/NUDIX hydrolase, [(2R,3S,5R)-5-(4-azanyl-5-methyl-pyrimidin-1-ium-1-yl)-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRC
 
 | | Structural insights into the catalysis mechanism of M. smegmatis antimutator protein MutT2 | | Descriptor: | 1,2-ETHANEDIOL, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRH
 
 | | M. smegmatis antimutator protein MutT2 in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRO
 
 | | M. smegmatis antimutator protein MutT2 in complex with 5mdCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5DYS
 
 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, PALMITIC ACID, RETINAL, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-09-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
5EN0
 
 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-3, PALMITIC ACID, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-11-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
4BEZ
 
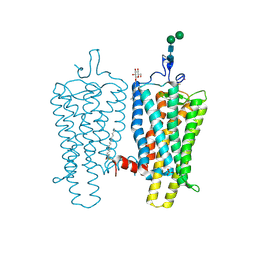 | | Night blindness causing G90D rhodopsin in the active conformation | | Descriptor: | ACETATE ION, PALMITIC ACID, RHODOPSIN, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
4BEY
 
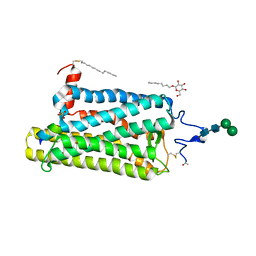 | | Night blindness causing G90D rhodopsin in complex with GaCT2 peptide | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
8GKF
 
 | |
5HBC
 
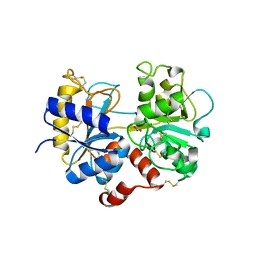 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
7MIE
 
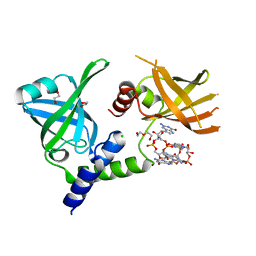 | | Crystal structure of the Borreliella burgdorferi PlzA protein in complex with c-di-GMP | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, ... | | Authors: | Davies, C, Singh, A. | | Deposit date: | 2021-04-16 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structure of the Borreliella burgdorferi PlzA protein in complex with c-di-GMP: new insights into the interaction of c-di-GMP with the novel xPilZ domain.
Pathog Dis, 79, 2021
|
|
4OQO
 
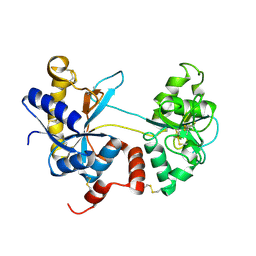 | | Crystal structure of the tryptic generated iron-free C-lobe of bovine Lactoferrin at 2.42 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lactotransferrin | | Authors: | Singh, A, Rastogi, N, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the iron-free true C-terminal half of bovine lactoferrin produced by tryptic digestion and its functional significance in the gut.
Febs J., 281, 2014
|
|
6LF7
 
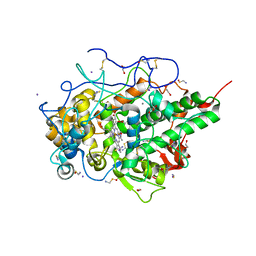 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Tyagi, T.K, Singh, R.P, Singh, A.K, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-30 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution.
To Be Published
|
|
8ZOZ
 
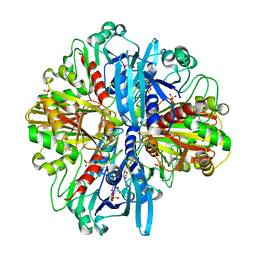 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
