5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
1D2U
 
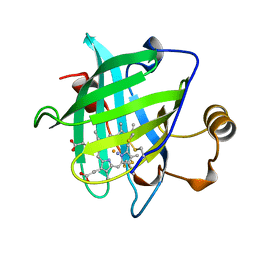 | | 1.15 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 1999-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
8Y2C
 
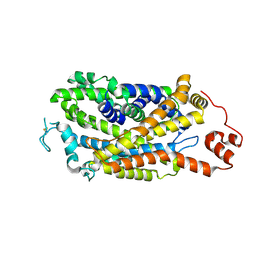 | | Cryo-EM structure of human dopamine transporter in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium-dependent dopamine transporter | | Authors: | Zhao, Y, Li, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Dopamine reuptake and inhibitory mechanisms in human dopamine transporter.
Nature, 632, 2024
|
|
8Y2E
 
 | |
8Y2G
 
 | |
8Y2F
 
 | | Cryo-EM structure of human dopamine transporter in complex with GBR12909 | | Descriptor: | 1-[2-[bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium-dependent dopamine transporter | | Authors: | Zhao, Y, Li, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Dopamine reuptake and inhibitory mechanisms in human dopamine transporter.
Nature, 632, 2024
|
|
8Y2D
 
 | |
9IUL
 
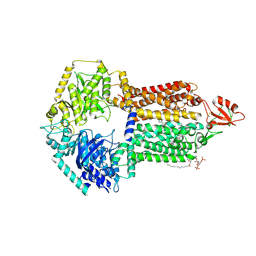 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUK
 
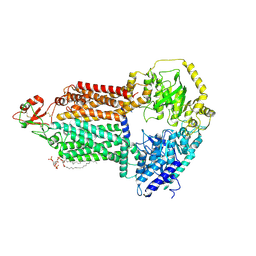 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUM
 
 | |
4NIE
 
 | |
8Y7L
 
 | |
8WBX
 
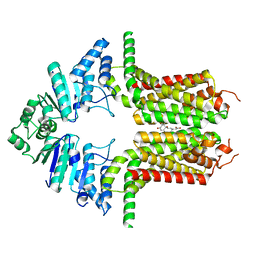 | | Cryo-EM structure of the ABCG25 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-10 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WD6
 
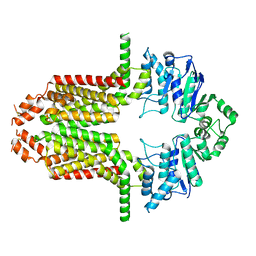 | | Cryo-EM structure of the ABCG25 | | Descriptor: | ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WAM
 
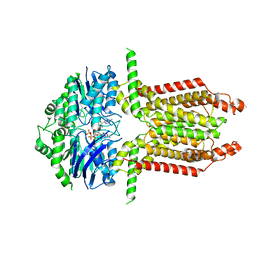 | |
8WBA
 
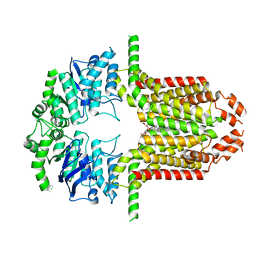 | | Cryo-EM structure of the ABCG25 bound to CHS | | Descriptor: | ABC transporter G family member 25, CHOLESTEROL HEMISUCCINATE | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-09 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
1IKJ
 
 | | 1.27 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH IMIDAZOLE | | Descriptor: | IMIDAZOLE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qui, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
4LCD
 
 | |
5ZNS
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
5ZNT
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GZ1
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8GZ2
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
6CRM
 
 | |
1ML7
 
 | | Crystal structure of nitrophorin 4 complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, nitrophorin 4 | | Authors: | Berry, R.E, Ding, X.D, Weichsel, A, Montfort, W.R, Walker, F.A. | | Deposit date: | 2002-08-30 | | Release date: | 2002-09-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Axial ligand complexes of the Rhodnius nitrophorins: reduction potentials, binding constants, EPR spectra, and structures of the 4-iodopyrazole and imidazole complexes of NP4
J.Biol.Inorg.Chem., 9, 2004
|
|
