7LEQ
 
 | |
7N9H
 
 | |
6RGV
 
 | |
3GZ1
 
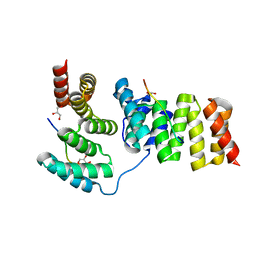 | | Crystal structure of IpgC in complex with the chaperone binding region of IpaB | | Descriptor: | Chaperone protein ipgC, GLYCEROL, Invasin ipaB | | Authors: | Lunelli, M, Lokareddy, R.K, Zychlinsky, A, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | IpaB-IpgC interaction defines binding motif for type III secretion translocator
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GYZ
 
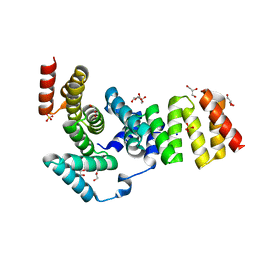 | | Crystal structure of IpgC from Shigella flexneri | | Descriptor: | Chaperone protein ipgC, GLYCEROL, SODIUM ION, ... | | Authors: | Lunelli, M, Lokareddy, R.K, Zychlinsky, A, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | IpaB-IpgC interaction defines binding motif for type III secretion translocator
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8DKR
 
 | |
8VXQ
 
 | |
6XMI
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI1
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI2
 
 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
9BGM
 
 | | Pseudomonas phage DEV neck and tail (portal, head-to-tail and tail tube proteins) | | Descriptor: | gp75 tail tube, gp80 portal protein, gp83 head-to-tail | | Authors: | Iglesias, S.M, Hou, C.-F.D, Li, F, Cingolani, G. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Integrative structural analysis of Pseudomonas phage DEV reveals a genome ejection motor.
Nat Commun, 15, 2024
|
|
9COD
 
 | | C15 symmetrized DEV collar | | Descriptor: | SGNH hydrolase-type esterase domain-containing protein | | Authors: | Iglesias, S.M, Hou, C.F.D, Li, F, Cingolani, G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Integrative structural analysis of Pseudomonas phage DEV reveals a genome ejection motor.
Nat Commun, 15, 2024
|
|
9BGO
 
 | |
9BGN
 
 | |
7JOQ
 
 | | Structure of NV1 small terminase | | Descriptor: | Small Terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
6E3B
 
 | |
6E6A
 
 | | Triclinic crystal form of IncA G144A point mutant | | Descriptor: | Inclusion membrane protein A, SODIUM ION | | Authors: | Cingolani, G, Paumet, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA.
Nat Commun, 10, 2019
|
|
6W7T
 
 | | Structure of PaP3 small terminase | | Descriptor: | small terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
7K5C
 
 | |
8G5V
 
 | | Empty capsid of Hepatitis B virus | | Descriptor: | Core protein Cp183 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2023-02-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
8G6V
 
 | |
8G8Y
 
 | |
8GCN
 
 | |
6E7E
 
 | |
8U11
 
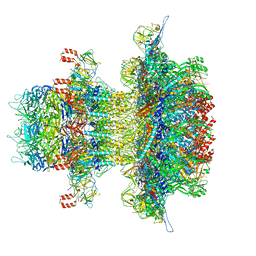 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
