3CEG
 
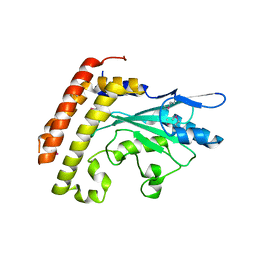 | | Crystal structure of the UBC domain of baculoviral IAP repeat-containing protein 6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
7MPS
 
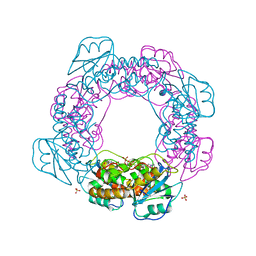 | | Brucella melitensis NrnC with engaged loop | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQB
 
 | | Bartonella henselae NrnC bound to pGG. D4 Symmetry | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQD
 
 | | Bartonella henselae NrnC complexed with pAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPL
 
 | | Bartonella henselae NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQG
 
 | | Bartonella henselae NrnC complexed with pAAAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPM
 
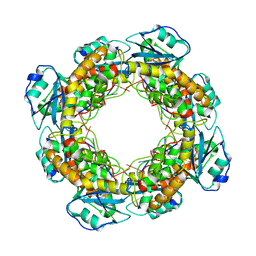 | | Bartonella henselae NrnC bound to pAA | | Descriptor: | 5'-phosphorylated AA, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPP
 
 | |
7MPN
 
 | | Bartonella henselae NrnC bound to pGC | | Descriptor: | 5'-phosphorylated GC, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPR
 
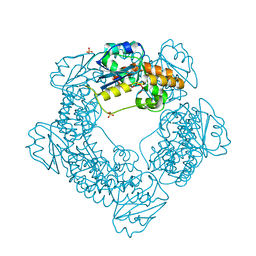 | | Brucella melitensis NrnC | | Descriptor: | NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQC
 
 | | Bartonella henselae NrnC bound to pGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPO
 
 | | Bartonella henselae NrnC bound to pAp | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQI
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPQ
 
 | |
7MQH
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. D4 Symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPT
 
 | | Brucella melitensis NrnC with bound Mg2+ | | Descriptor: | MAGNESIUM ION, NanoRNase C, PHOSPHATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQF
 
 | | Bartonella henselae NrnC complexed with pAAAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPU
 
 | | Brucella melitensis NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQE
 
 | | Bartonella henselae NrnC complexed with pAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
1UKH
 
 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, Mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
1UKI
 
 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
5D4Z
 
 | |
5D50
 
 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1P1P
 
 | | [PRO7,13] AA-CONOTOXIN PIVA, NMR, 12 STRUCTURES | | Descriptor: | AA-CONOTOXIN PIVA | | Authors: | Han, K.-H, Hwang, K.-J, Kim, S.-M, Kim, S.-K, Gray, W.R, Olivera, B.M, Rivier, J, Shon, K.J. | | Deposit date: | 1996-12-06 | | Release date: | 1997-07-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a novel conotoxin, [Pro 7,13] alpha A-conotoxin PIVA.
Biochemistry, 36, 1997
|
|
5YSO
 
 | | Crystal structure of Estrogen Related Receptor-3 (ERR-gamma) ligand binding domain with DN200434 | | Descriptor: | 4-[5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Cho, S.J, Chin, J.W, Yoon, H.S, Jeon, Y.H, Bae, J.H, Song, J.Y. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | A Novel Orally Active Inverse Agonist of Estrogen-related Receptor Gamma (ERR gamma ), DN200434, A Booster of NIS in Anaplastic Thyroid Cancer.
Clin.Cancer Res., 25, 2019
|
|
