4P31
 
 | | Crystal structure of a selenomethionine derivative of E. coli LptB in complex with ADP-Magensium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RH8
 
 | |
4RHB
 
 | |
1DCM
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N WITH AN ATYPICAL CONFORMER (MONOMER A) | | Descriptor: | MAGNESIUM ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DBW
 
 | | CRYSTAL STRUCTURE OF FIXJ-N | | Descriptor: | POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DCK
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N COMPLEXED WITH MN2+ | | Descriptor: | MANGANESE (II) ION, POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1D5W
 
 | | PHOSPHORYLATED FIXJ RECEIVER DOMAIN | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Birck, C, Mourey, L, Gouet, P, Fabry, B, Schumacher, J, Rousseau, P, Kahn, D, Samama, J.P. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes induced by phosphorylation of the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
6B8B
 
 | | E. coli LptB in complex with ADP and a novobiocin derivative | | Descriptor: | (3s,5s,7s)-N-{7-[(3-O-carbamoyl-6-deoxy-5-methyl-4-O-methyl-beta-D-gulopyranosyl)oxy]-4-hydroxy-8-methyl-2-oxo-2H-1-ben zopyran-3-yl}tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Mandler, M.D, Owens, T.W, Lazarus, M.B, May, J.M, Kahne, D.K. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
6B89
 
 | | E. coli LptB in complex with ADP and novobiocin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION, ... | | Authors: | May, J.M, Lazarus, M.B, Sherman, D.J, Owens, T.W, Mandler, M.D, Kahne, D.K. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
3NB7
 
 | |
2OQO
 
 | |
3D3H
 
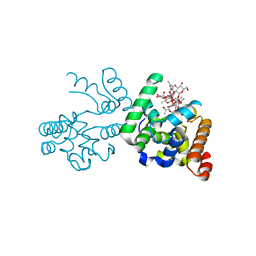 | | Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A | | Descriptor: | (2R)-3-{[(S)-{[(2R,3R,4R,5S,6S)-3-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2R,3R,4S,5R,6S)-6-carbamoyl-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}methyl)tetrahydro-2H-pyran-2-yl]oxy}-6-carbamoyl-4-(carbamoyloxy)-5-hydroxy-5-methyltetrahydro-2H-pyran-2-yl]oxy}(hydroxy)phosphoryl]oxy}-2-{[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]oxy}propanoic acid, Penicillin-insensitive transglycosylase | | Authors: | Yuan, Y, Sliz, P, Walker, S. | | Deposit date: | 2008-05-11 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design.
Acs Chem.Biol., 3, 2008
|
|
2GV2
 
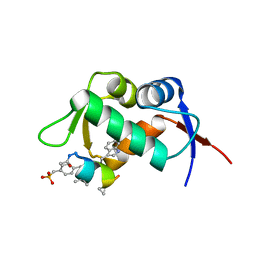 | | MDM2 in complex with an 8-mer p53 peptide analogue | | Descriptor: | 8-MER P53 PEPTIDE ANALOGUE, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Schubert, C, Sakurai, K. | | Deposit date: | 2006-05-02 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Analysis of an 8-mer p53 Peptide Analogue Complexed with MDM2.
J.Am.Chem.Soc., 128, 2006
|
|
6PL6
 
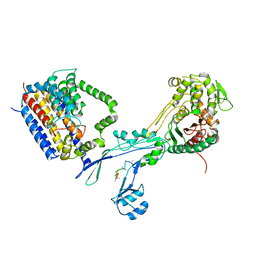 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAS
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6MGF
 
 | | untagged, wild-type LptB in complex with ADP | | Descriptor: | (3s,5s,7s)-N-{7-[(3-O-carbamoyl-6-deoxy-5-methyl-4-O-methyl-beta-D-gulopyranosyl)oxy]-4-hydroxy-8-methyl-2-oxo-2H-1-ben zopyran-3-yl}tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Mandler, M.D, Owens, T.W, Ruiz, N, Kahne, D. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Bypassing the requirement for coupling of ATP binding and hydrolysis in the lipopolysaccharide ABC transporte
not published
|
|
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
