4I9Q
 
 | | Crystal structure of the ternary complex of the D714A mutant of RB69 DNA polymerase | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), ... | | Authors: | Guja, K.E, Jacewicz, A, Trzemecka, A, Plochocka, D, Yakubovskaya, E, Bebenek, A, Garcia-Diaz, M. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Remote Palm Domain Residue of RB69 DNA Polymerase Is Critical for Enzyme Activity and Influences the Conformation of the Active Site.
Plos One, 8, 2013
|
|
4KHN
 
 | | Crystal structure of the ternary complex of the D714A mutant of RB69 DNA polymerase | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), ... | | Authors: | Guja, K.E, Jacewicz, A, Trzemecka, A, Plochocka, D, Yakubovskaya, E, Bebenek, A, Garcia-Diaz, M. | | Deposit date: | 2013-04-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Remote Palm Domain Residue of RB69 DNA Polymerase Is Critical for Enzyme Activity and Influences the Conformation of the Active Site.
Plos One, 8, 2013
|
|
4IP2
 
 | | Putative Aromatic Acid Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, Aromatic Acid Decarboxylase, GLYCEROL, ... | | Authors: | Schneider, G, Brunner, K, Izumi, A, Jacewicz, A. | | Deposit date: | 2013-01-09 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the UbiD protein family from the crystal structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
7T7N
 
 | | Structure of SPCC1393.13 protein from fission yeast | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Damage-control phosphatase SPCC1393.13, PHOSPHATE ION | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7T7O
 
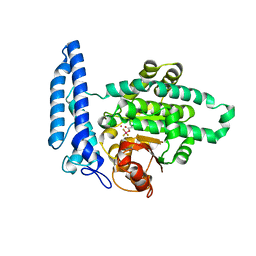 | |
7T7K
 
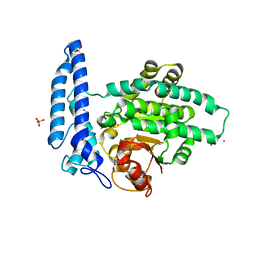 | | Structure of SPAC806.04c protein from fission yeast bound to Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Damage-control phosphatase SPAC806.04c, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7U1Y
 
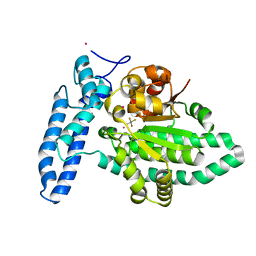 | | Structure of SPAC806.04c protein from fission yeast bound to AlF4 and Co2+ | | Descriptor: | COBALT (II) ION, Damage-control phosphatase SPAC806.04c, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7U1V
 
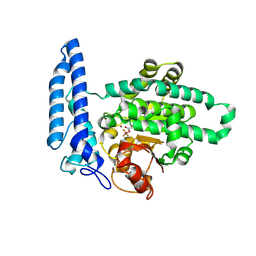 | | Structure of SPAC806.04c protein from fission yeast covalently bound to BeF3 | | Descriptor: | Damage-control phosphatase SPAC806.04c, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7U1X
 
 | |
5V9X
 
 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | Descriptor: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | Deposit date: | 2017-03-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
6XQN
 
 | | Structure of a mitochondrial calcium uniporter holocomplex (MICU1, MICU2, MCU, EMRE) in low Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, Calcium uptake protein 1, ... | | Authors: | Long, S.B, Wang, C, Baradaran, R, Jacewicz, A, Delgado, B. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures reveal gatekeeping of the mitochondrial Ca 2+ uniporter by MICU1-MICU2.
Elife, 9, 2020
|
|
6XQO
 
 | | Structure of the human MICU1-MICU2 heterodimer, calcium bound, in association with a lipid nanodisc | | Descriptor: | CALCIUM ION, Calcium uptake protein 1, mitochondrial, ... | | Authors: | Long, S.B, Wang, C, Baradaran, R, Jacewicz, A, Delgado, B. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures reveal gatekeeping of the mitochondrial Ca 2+ uniporter by MICU1-MICU2.
Elife, 9, 2020
|
|
4MDF
 
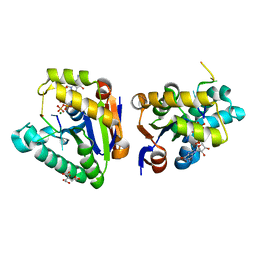 | | Structure of bacterial polynucleotide kinase Michaelis complex bound to GTP and DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
4MDE
 
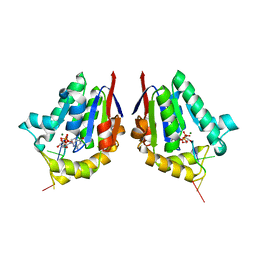 | | Structure of bacterial polynucleotide kinase product complex bound to GDP and DNA | | Descriptor: | DNA (5'-D(P*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
2XU8
 
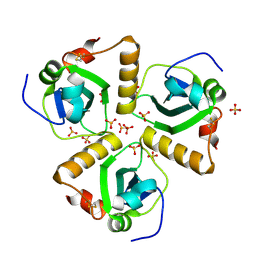 | | Structure of Pa1645 | | Descriptor: | PA1645, SULFATE ION | | Authors: | Abdelli, W.B, Moynie, L, McMahon, S.A, Liu, H, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2010-10-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ETR
 
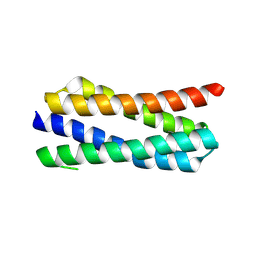 | | X-ray structure of PA2169 from Pseudomonas aeruginosa | | Descriptor: | Putative uncharacterized protein | | Authors: | Schnell, R, Sandalova, T, Lindqvist, Y, Schneider, G. | | Deposit date: | 2012-04-24 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4EXB
 
 | |
4ES6
 
 | |
4EXA
 
 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B9A
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B79
 
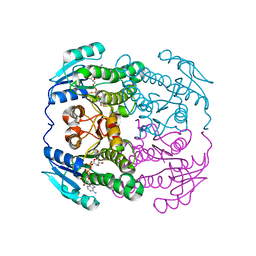 | | THE AEROPATH PROJECT AND PSEUDOMONAS AERUGINOSA HIGH-THROUGHPUT CRYSTALLOGRAPHIC STUDIES FOR ASSESSMENT OF POTENTIAL TARGETS IN EARLY STAGE DRUG DISCOVERY. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B7C
 
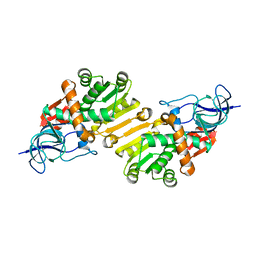 | | Crystal structure of hypothetical protein PA1648 from Pseudomonas aeruginosa. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PROBABLE OXIDOREDUCTASE | | Authors: | Alphey, M.S, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-25 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B7X
 
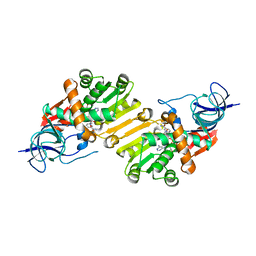 | | Crystal structure of hypothetical protein PA1648 from Pseudomonas aeruginosa. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROBABLE OXIDOREDUCTASE | | Authors: | Alphey, M.S, McMahon, S.A, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AVY
 
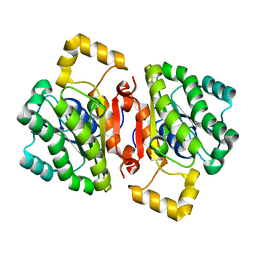 | | The AEROPATH project and Pseudomonas aeruginosa high-throughput crystallographic studies for assessment of potential targets in early stage drug discovery. | | Descriptor: | PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
