5ZSJ
 
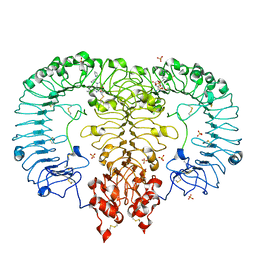 | | Crystal structure of monkey TLR7 in complex with GS9620 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-azanyl-2-butoxy-8-[[3-(pyrrolidin-1-ylmethyl)phenyl]methyl]-5,7-dihydropteridin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSG
 
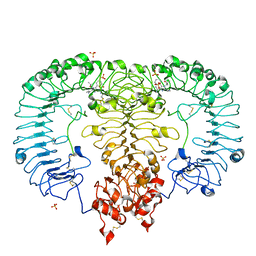 | | Crystal structure of monkey TLR7 in complex with gardiquimod | | Descriptor: | 1-{4-amino-2-[(ethylamino)methyl]-1H-imidazo[4,5-c]quinolin-1-yl}-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
7SS5
 
 | |
5ZSN
 
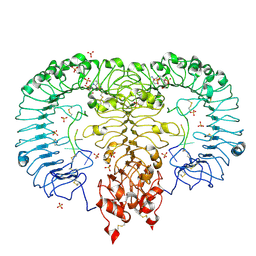 | | Crystal structure of monkey TLR7 in complex with AAUUAA | | Descriptor: | 2',3'- cyclic AMP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSI
 
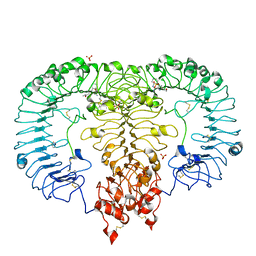 | | Crystal structure of monkey TLR7 in complex with CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSC
 
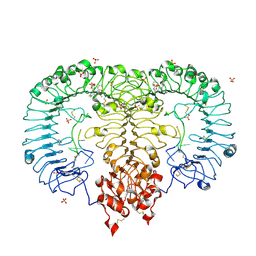 | | Crystal structure of monkey TLR7 in complex with IMDQ and CCUUCC | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
8HD0
 
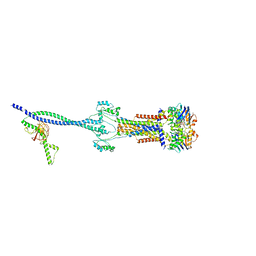 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Chen, Y. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insight into the septal peptidoglycan hydrolysis machinery of bacterial cell division
To Be Published
|
|
6JIO
 
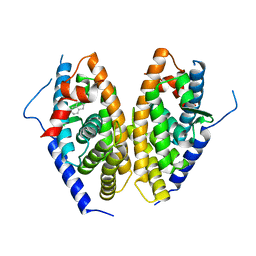 | | Human LXR-beta in complex with a ligand | | Descriptor: | Oxysterols receptor LXR-beta, tert-butyl 7-amino-3,4-dihydroisoquinoline-2(1H)-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identify liver X receptor beta modulator building blocks by developing a fluorescence polarization-based competition assay.
Eur.J.Med.Chem., 178, 2019
|
|
2YU2
 
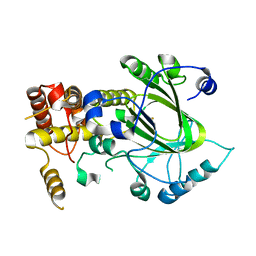 | | Crystal structure of hJHDM1A without a-ketoglutarate | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
2YU1
 
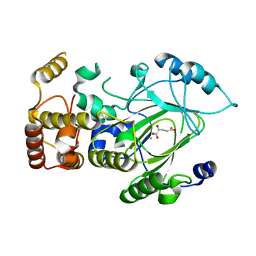 | | Crystal structure of hJHDM1A complexed with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
2QXV
 
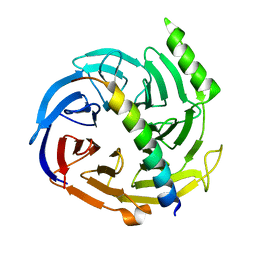 | | Structural basis of EZH2 recognition by EED | | Descriptor: | Embryonic ectoderm development, Enhancer of zeste homolog 2 | | Authors: | Han, Z. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of EZH2 recognition by EED
Structure, 15, 2007
|
|
4D4Z
 
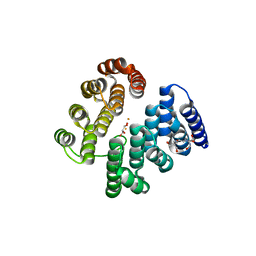 | | STRUCTURE OF HUMAN DEOXYHYPUSINE HYDROXYLASE in complex with glycerol | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYHYPUSINE HYDROXYLASE, FE (III) ION, ... | | Authors: | Han, Z, Sakai, N, Hilgenfeld, R. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Peroxo-Diiron(III) Intermediate of Deoxyhypusine Hydroxylase, an Oxygenase Involved in Hypusination.
Structure, 23, 2015
|
|
4D50
 
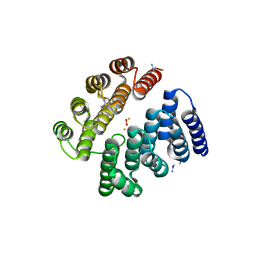 | | Structure of human deoxyhypusine hydroxylase | | Descriptor: | DEOXYHYPUSINE HYDROXYLASE, FE (III) ION, GUANIDINE, ... | | Authors: | Han, Z, Sakai, N, Hilgenfeld, R. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Peroxo-Diiron(III) Intermediate of Deoxyhypusine Hydroxylase, an Oxygenase Involved in Hypusination.
Structure, 23, 2015
|
|
8I6I
 
 | |
7OQE
 
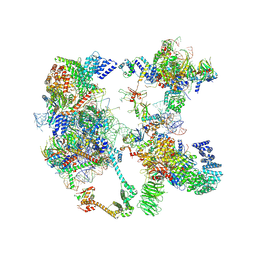 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQB
 
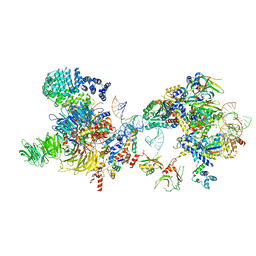 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQC
 
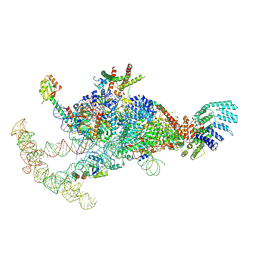 | | The U1 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A), Pre-mRNA-processing factor 39, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
5LSQ
 
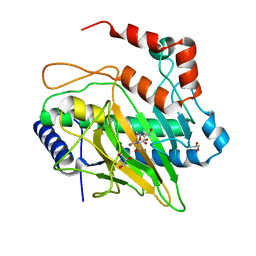 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ethylene Forming Enzyme, ... | | Authors: | Zhang, Z, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3FKD
 
 | |
3EVZ
 
 | |
3GG4
 
 | |
3GBT
 
 | |
3DZB
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
3G1W
 
 | |
