6TV0
 
 | | Serratia spp. cyanase hydratase | | Descriptor: | Cyanate hydratase, GLYCEROL, OXALIC ACID | | Authors: | Pederzoli, R, Tarantino, D, Gourlay, L.J, Chaves-Sanjuan, A, Bolognesi, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detecting the nature and solving the crystal structure of a contaminant protein from an opportunistic pathogen.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5N2C
 
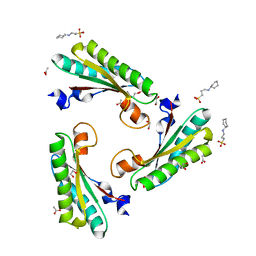 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, ... | | Authors: | Matterazzo, E, Bolognesi, M, Gourlay, L.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing Probes for Immunodiagnostics: Structural Insights into an Epitope Targeting Burkholderia Infections.
ACS Infect Dis, 3, 2017
|
|
4UU4
 
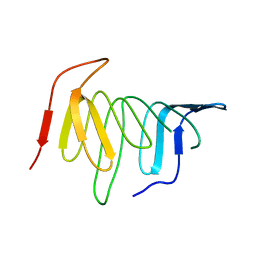 | | Crystal structure of LptH, the LptA homologous periplasmic component of the conserved lipopolysaccharide transport device from Pseudomonas aeruginosa | | Descriptor: | PERIPLASMIC LIPOPOLYSACCHARIDE TRANSPORT PROTEIN LPTH | | Authors: | Bollati, M, Villa, R, Gourlay, L.J, Barbiroli, A, Deho, G, Benedet, M, Polissi, A, Martorana, A, Sperandeo, P, Bolognesi, M, Nardini, M. | | Deposit date: | 2014-07-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal Structure of Lpth, the Periplasmic Component of the Lipopolysaccharide Transport Machinery from Pseudomonas Aeruginosa.
FEBS J., 282, 2015
|
|
3ZS6
 
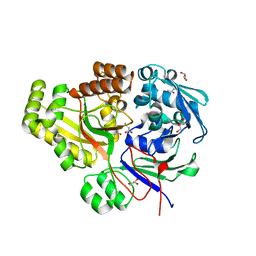 | | The Structural characterization of Burkholderia pseudomallei OppA. | | Descriptor: | CHLORIDE ION, GLYCEROL, OLIGOPEPTIDE DVA, ... | | Authors: | Lassaux, P, Gourlay, L.J, Bolognesi, M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structure-Based Strategy for Epitope Discovery in Burkholderia Pseudomallei Oppa Antigen.
Structure, 21, 2013
|
|
6Y0D
 
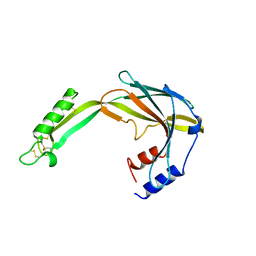 | |
2C80
 
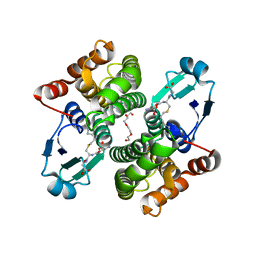 | | Structure of Sh28GST in complex with S-hexyl Glutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE 28 KDA, S-HEXYLGLUTATHIONE, TETRAETHYLENE GLYCOL | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Brunori, M, Miele, A.E. | | Deposit date: | 2005-11-30 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CAI
 
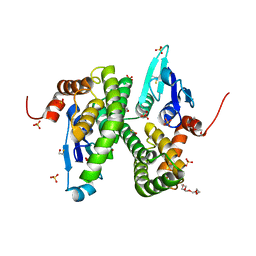 | | Structure of Glutathione-S-Transferase mutant, R21L, from Schistosoma Haematobium | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE S-TRANSFERASE 28 KDA, SULFATE ION, ... | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Brunori, M, Miele, A.E. | | Deposit date: | 2005-12-21 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2C8U
 
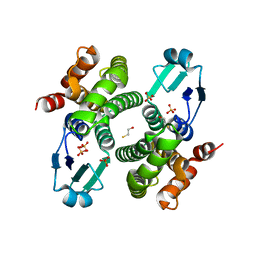 | | Structure of R21Q mutant of Sh28GST | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE S-TRANSFERASE 28 KDA, SULFATE ION | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-07 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CA8
 
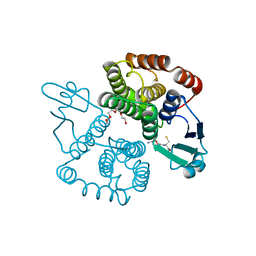 | | Structure of Sh28GST in complex with GSH at pH 6.0 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE 28 KDA, TETRAETHYLENE GLYCOL | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CAQ
 
 | | Structure of R21L mutant of Sh28GST in complex with GSH | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE 28 KDA, ... | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
4CFI
 
 | | 3D structure of FliC from Burkholderia pseudomallei | | Descriptor: | FLAGELLIN | | Authors: | Lassaux, P, Peri, C, Ferrer-Navarro, M, Gourlay, L.J, Conchillo-Sole, O, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sequence- and Structure-Based Immunoreactive Epitope Discovery for Burkholderia Pseudomallei Flagellin.
Plos Negl Trop Dis, 9, 2015
|
|
4B54
 
 | | The Structure of the inactive mutant G153R of LptC from E. coli | | Descriptor: | ACETATE ION, LIPOPOLYSACCHARIDE EXPORT SYSTEM PROTEIN LPTC | | Authors: | Villa, R, Martorana, A.M, Sperandeo, P, Kahne, D, Okuda, S, Gourlay, L.J, Nardini, M, Bolognesi, M, Polissi, A. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Escherichia Coli Lpt Transenvelope Protein Complex for Lipopolysaccharide Export is Assembled Via Conserved Structurally Homologous Domains.
J.Bacteriol., 195, 2013
|
|
6YN7
 
 | |
7OFN
 
 | |
6SSV
 
 | | The structure of serpin from Schistosoma mansoni | | Descriptor: | Serpin, putative | | Authors: | De Benedetti, S, Gourlay, L. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application.
Vaccines (Basel), 9, 2021
|
|
4BYZ
 
 | | Structural characterization using Sulfur-SAD of the cytoplasmic domain of Burkholderia pseudomallei PilO2Bp, an actin-like protein component of a Type IVb R64-derivative pilus machinery. | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, TYPE IV PILUS BIOSYNTHESIS PROTEIN | | Authors: | Lassaux, P, Manjasetty, B.A, Conchillo-Sole, O, Yero, D, Gourlay, L, Perletti, L, Daura, X, Belrhali, H, Bolognesi, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Redefining the Pf06864 Pfam Family Based on Burkholderia Pseudomallei Pilo2BP S-Sad Crystal Structure.
Plos One, 9, 2014
|
|
4BZ0
 
 | | Structural characterization using Sulfur-SAD of the cytoplasmic domain of Burkholderia pseudomallei PilO2Bp, an actin-like protein component of a Type IVb R64-derivative pilus machinery. | | Descriptor: | POTASSIUM ION, PUTATIVE TYPE IV PILUS BIOSYNTHESIS PROTEIN | | Authors: | Lassaux, P, Manjasetty, B.A, Conchillo-Sole, O, Yero, D, Gourlay, L, Perletti, L, Daura, X, Belrhali, H, Bolognesi, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Redefining the Pf06864 Pfam Family Based on Burkholderia Pseudomallei Pilo2BP S-Sad Crystal Structure.
Plos One, 9, 2014
|
|
