6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
9I8M
 
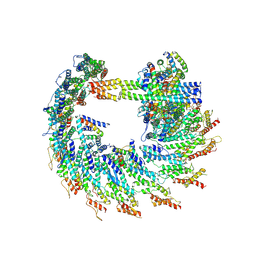 | |
9I8N
 
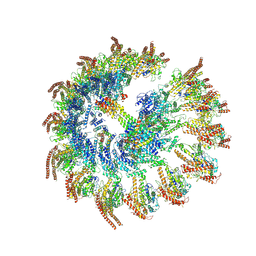 | |
9I8H
 
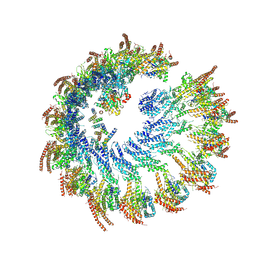 | |
9I8G
 
 | |
4IAN
 
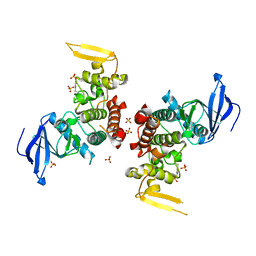 | | Crystal Structure of apo Human PRPF4B kinase domain | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-06 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IFC
 
 | | Crystal Structure of ADP-bound Human PRPF4B kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-14 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IJP
 
 | | Crystal Structure of Human PRPF4B kinase domain in complex with 4-{5-[(2-Chloro-pyridin-4-ylmethyl)-carbamoyl]-thiophen-2-yl}-benzo[b]thiophene-2-carboxylic acid amine | | Descriptor: | 4-(5-{[(2-chloropyridin-4-yl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IIR
 
 | | Crystal Structure of AMPPNP-bound Human PRPF4B kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
7CL8
 
 | |
5PZL
 
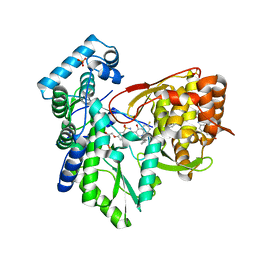 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 2-({3-[1-(2-CYCLOPROPYLETHYL)-6-FLUORO-4-HYDROXY-2-OXO-1,2-DIHYDROQUINOLIN-3-YL]-1,1-DIOXO-1,4-DIHYDRO-1LAMBDA~6~,2,4-BENZOTHIADIAZIN-7-YL}OXY)ACETAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-({3-[1-(2-cyclopropylethyl)-6-fluoro-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxo-1,4-dihydro-1lambda~6~,2,4-benzothiadiazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
5PZK
 
 | |
5PZP
 
 | |
5PZM
 
 | |
5PZN
 
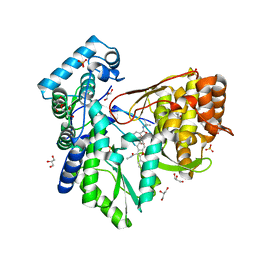 | |
5PZO
 
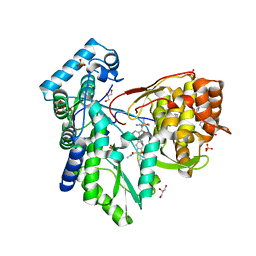 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE C316N IN COMPLEX WITH 2-(4-FLUOROPHENYL)-N-METHYL-5-[3-({2-METHYL-1-OXO-1-[(1,3,4-THIADIAZOL-2-YL)AMINO]PROPAN-2-YL}CARBAMOYL)PHENYL]-1-BENZOFURAN-3-CARBOXAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-(4-fluorophenyl)-N-methyl-5-[3-({2-methyl-1-oxo-1-[(1,3,4-thiadiazol-2-yl)amino]propan-2-yl}carbamoyl)phenyl]-1-benzofuran-3-carboxamide, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
9EOK
 
 | |
4UMX
 
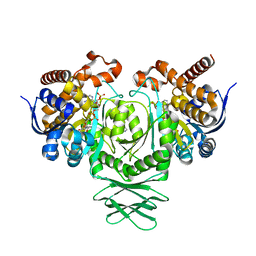 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
4UMY
 
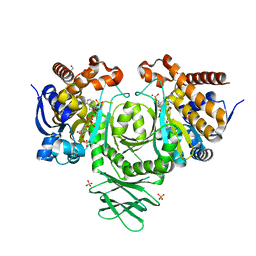 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McLean, L, Zhang, Y, Mathieu, M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
8Q34
 
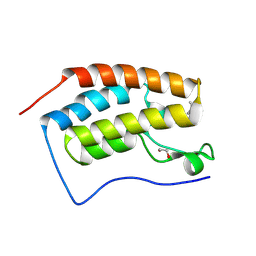 | | Crystal structure of the first bromodomain of human BRD4 in complex with the ligand ZZ001229a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1~{H}-imidazo[4,5-b]pyridin-2-ylmethyl)-3-(3-methyl-1,2-diazirin-3-yl)propanamide | | Authors: | MacLean, E.M, Gao, Q, Williams, E, Balcomb, B.H, von Delft, F, Bajusz, D, Keeley, A, Abranyi-Balogh, P, Koekemoer, L, Keseru, G.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein binding sites by photoreactive fragment pharmacophores.
Commun Chem, 7, 2024
|
|
1C9Z
 
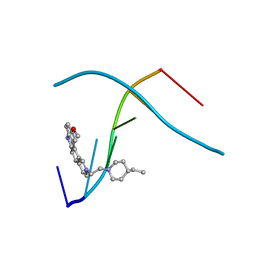 | | D232-CGTACG | | Descriptor: | 1,3-DI[[[10-METHOXY-7H-PYRIDO[4,3-C]CARBAZOL-2-IUMYL]-ETHYL]-PIPERIDIN-4-YL]-PROPANE, 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Williams, L.D. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of cationic charge on three-dimensional structures of intercalative complexes: structure of a bis-intercalated DNA complex solved by MAD phasing.
Curr.Med.Chem., 7, 2000
|
|
4NLD
 
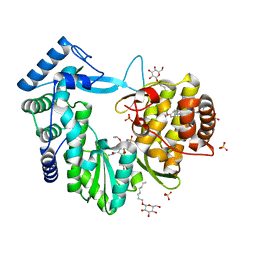 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
