4I8J
 
 | | Bovine trypsin at 0.87 A resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8L
 
 | | Bovine trypsin at 0.87 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BLU
 
 | | STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CHROMATIUM VINOSUM | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the 2[4Fe-4S] ferredoxin from Chromatium vinosum: evolutionary and mechanistic inferences for [3/4Fe-4S] ferredoxins.
Protein Sci., 5, 1996
|
|
1DUP
 
 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
1EGP
 
 | |
2VB1
 
 | | HEWL at 0.65 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LYSOZYME C, ... | | Authors: | Wang, J, Dauter, M, Alkire, R, Joachimiak, A, Dauter, Z. | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.65 Å) | | Cite: | Triclinic Lysozyme at 0.65 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2ID8
 
 | | Crystal structure of Proteinase K | | Descriptor: | (S)-(2,3-DIHYDROXYPROPOXY)TRIHYDROXYBORATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, J, Dauter, M, Dauter, Z. | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | What can be done with a good crystal and an accurate beamline?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
3CYR
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774P | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | Deposit date: | 1997-07-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the Three-Dimensional Structures of Cytochrome C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstroms Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstroms Resolution
Inorg.Chim.Acta., 273, 1998
|
|
4OCB
 
 | |
5MU4
 
 | | Tail Tubular Protein A of Klebsiella pneumoniae bacteriophage KP32 | | Descriptor: | Tail tubular protein A | | Authors: | Pyra, A, Brzozowska, E, Pawlik, K, Dauter, M, Dauter, Z, Gamian, A. | | Deposit date: | 2017-01-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tail tubular protein A: a dual-function tail protein of Klebsiella pneumoniae bacteriophage KP32.
Sci Rep, 7, 2017
|
|
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
3US6
 
 | | Crystal Structure of Histidine-containing Phosphotransfer Protein MtHPt1 from Medicago truncatula | | Descriptor: | Histidine-containing Phosphotransfer Protein type 1, MtHPt1 | | Authors: | Ruszkowski, M, Brzezinski, K, Jedrzejczak, R, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Medicago truncatula histidine-containing phosphotransfer protein: Structural and biochemical insights into the cytokinin transduction pathway in plants.
Febs J., 280, 2013
|
|
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
4GS3
 
 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HP4
 
 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3SIX
 
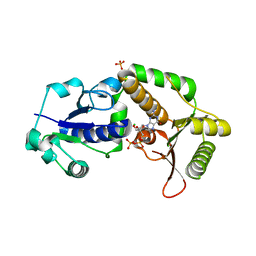 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SIW
 
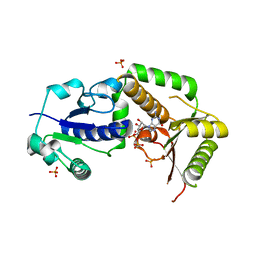 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2BAX
 
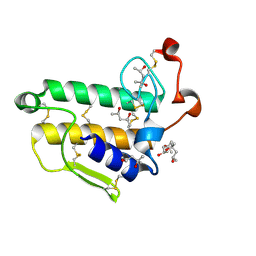 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
9EWK
 
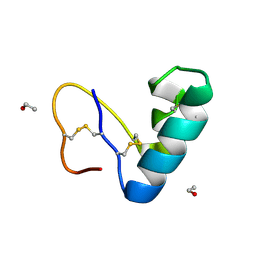 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
2F6D
 
 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
6M9C
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Pseudotyrostatin | | Descriptor: | ACETIC ACID, CALCIUM ION, Pseudotyrostatin, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6O64
 
 | |
6O63
 
 | |
3P4J
 
 | | Ultra-high resolution structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Brzezinski, K, Brzuszkiewicz, A, Dauter, M, Kubicki, M, Jaskolski, M, Dauter, Z. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.55 Å) | | Cite: | High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 A.
Nucleic Acids Res., 39, 2011
|
|
