1FYW
 
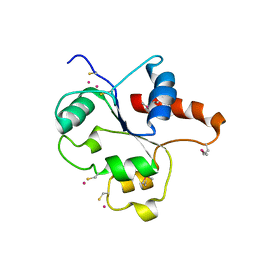 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
1HW2
 
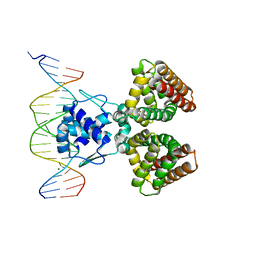 | | FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ECHERICHIA COLI | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*GP*TP*CP*CP*GP*AP*CP*CP*AP*GP*AP*TP*GP*CP*T)-3', 5'-D(*G*CP*AP*TP*CP*TP*GP*GP*TP*CP*GP*GP*AP*CP*CP*AP*GP*AP*TP*CP*GP*A)-3', FATTY ACID METABOLISM REGULATOR PROTEIN, ... | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1HW1
 
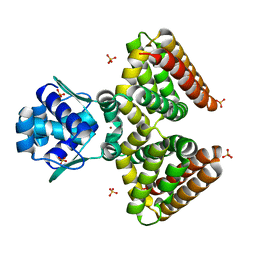 | | THE FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ESCHERICHIA COLI | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION, ZINC ION | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
5B7P
 
 | |
5B7G
 
 | |
5B7Q
 
 | |
5CB0
 
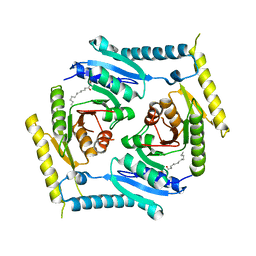 | | Crystal structure and functional implications of the tandem-type universal stress protein UspE from Escherichia coli | | Descriptor: | 3-oxotetradecanoic acid, Universal stress protein E | | Authors: | Xu, Y, Quan, C.S, Jin, X, Jin, L, Kim, J.S, Guo, J, Fan, S, Ha, N.C. | | Deposit date: | 2015-06-30 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Crystal structure and functional implications of the tandem-type universal stress protein UspE from Escherichia coli.
Bmc Struct.Biol., 16, 2016
|
|
2EEP
 
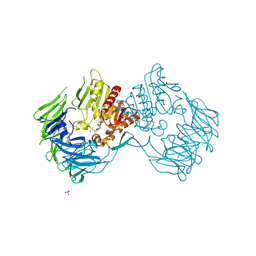 | | Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, putative, SULFATE ION, ... | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-02-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
6VWI
 
 | | Head region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
5DNW
 
 | | Crystal structure of KAI2-like protein from Striga (apo state 1) | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNU
 
 | | Crystal structure of Striga KAI2-like protein in complex with karrikin | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2H-furo[2,3-c]pyran-2-one, BENZOIC ACID, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
6VWH
 
 | |
5B7N
 
 | |
6VWG
 
 | | Head region of the open conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
6VWJ
 
 | |
2H25
 
 | |
4DK0
 
 | |
1QY7
 
 | | The structure of the PII protein from the cyanobacteria Synechococcus sp. PCC 7942 | | Descriptor: | NICKEL (II) ION, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QR6
 
 | | HUMAN MITOCHONDRIAL NAD(P)-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xu, Y, Bhargava, G, Wu, H, Loeber, G, Tong, L. | | Deposit date: | 1999-06-18 | | Release date: | 1999-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human mitochondrial NAD(P)(+)-dependent malic enzyme: a new class of oxidative decarboxylases.
Structure, 7, 1999
|
|
4DK1
 
 | |
1WDF
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
1WDG
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
1WP7
 
 | | crystal structure of Nipah Virus fusion core | | Descriptor: | fusion protein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nipah Virus fusion core
To be Published
|
|
1WNC
 
 | | Crystal structure of the SARS-CoV Spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Lou, Z, Liu, Y, Pang, H, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
J.Biol.Chem., 279, 2004
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
